FIGURE 9.
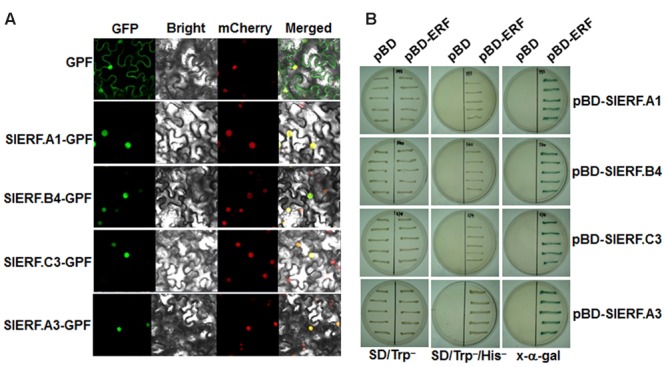
Subcellular localization and transactivation activity of SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 proteins. (A) Subcellular localization of SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 proteins. Agrobacteria carrying pFGC-Egfp-SlERF.A1, pFGC-Egfp-SlERF.A3, pFGC-Egfp-SlERF.B4, pFGC-Egfp-SlERF.C3, or pFGC-Egfp were infiltrated into leaves of N. benthamiana plants expressing a red nucleus marker RFP-H2B protein and the images were taken at 36 h after infiltration under dark field for green fluorescence (left) and red fluorescence (middle right), white field for cell morphology (middle left) and in combination (right), respectively. (B) Transactivation activity of SlERF.A1, SlERF.B4, and SlERF.C3 proteins in yeast. Yeast cells transformed with pBD-SlERF.A1, pBD-SlERF.A3, pBD-SlERF.B4, pBD-SlERF.C3 or pBD empty vector (as a negative control) were streaked on SD/Trp- plates (left) or SD/Trp-His- plates (middle) for 3 days at 28°C. The x-α-gal was added to the SD/Trp-His- plates and kept at 28°C for 6 hr (right).
