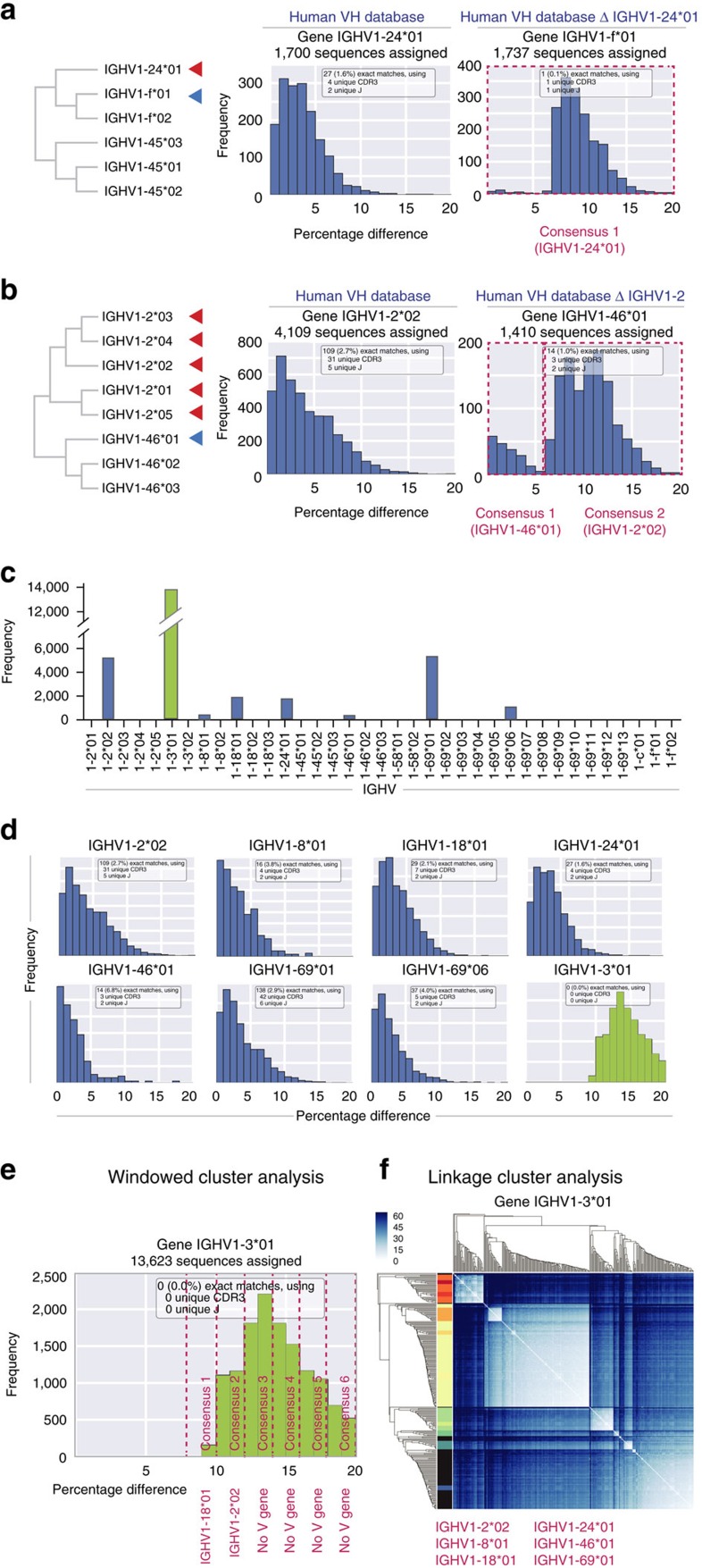
Figure 2. Identification of human VH alleles when the human reference database is rendered incomplete.
Phylogenetic tree of relationship between (a) IGHV1-24*01 and IGHV1-f*01 or (b) the IGHV1-2 alleles and IGHV1-46*01, where the red arrow shows the deleted allele and the blue arrow shows the closest allele to which the sequences are now assigned. Frequency plot of sequences assigned to (a) IGHV1-24*01 (centre panel) or (b) IGHV1-2*02 (centre panel) when the reference database is complete. The right panels shows the new assignment when IGHV1-24*01 (a) or IGHV1-2 alleles (b) are deleted from the reference database. The pink text below the right panels indicate the identification of one consensus sequence from the 1–20% range, identical to IGHV1-24*01 (a) or two consensus sequences from 1–6% and 6–20%, showing 100% identity to IGHV1-46*01 and IGHV1-2*02, respectively (b). (c) Bar chart showing the assignment of IGHV1 human IgM sequences using the complete starting database (blue bars) or an incomplete database from which all IGHV1 alleles apart from IGHV1-3*01 have been removed (green bars). (d) Frequency plots of the seven IGHV1 alleles expressed in the human IgM library (blue histograms) and the resultant frequency plot after all IGHV1 alleles apart from IGHV1-3*01 have been removed (green histogram). (e) Frequency plot of sequences assigned to IGHV1-3*01 showing Windowed cluster analysis. The assigned sequences are binned in 2% intervals and from each a consensus sequence is built. The pink text beneath the panel reveals the identification of consensus sequences, from the 8–10% and the 10–12% windows, showing 100% identity to IGHV1-18*01 and IGHV1-2*02, respectively. (f) Matrix diagram showing Linkage cluster analysis of sequences assigned to IGHV1-3*01 following deletion of the other IGHV1 alleles from the VH screening database. Clusters of closely related sequences are visualized as lighter coloured squares within a darker blue background. The coloured bars on the left of the figure show the numbers of clusters identified in this analysis. The red text beneath the panel reveals that consensus sequences from the linkage clusters were identical to six of the seven IGHV1 sequences.