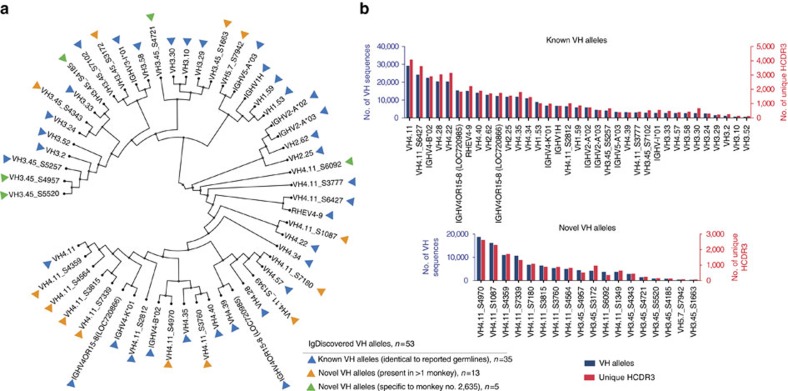
Figure 6. Analysis of novel VH sequences identified by IgDiscover in an Indian-origin rhesus macaque.
(a) Phylogenetic tree of VH alleles (n=53) from rhesus macaque no. 2635, analysed using a minimal seven gene starting database (VH1.53, VH2.12, VH3.45, VH4.11, VH5.7, VH6.1 and VH7.21). The resulting germline sequences show 100% identity to rhesus genomic reference sequence or to previously published germline alleles (blue arrows, n=35), 100% identity to germline alleles present in additional Indian rhesus animals (orange arrows, n=13) or present only in rhesus macaque no. 2635 (green arrows, n=5). (b) Frequency of the usage of novel alleles. Panels showing the results of V sequence assignment of rhesus macaque no. 2635 using the database created by IgDiscover. The blue columns represent the number of sequences assigned to the database allele, while the red columns represent the numbers of unique CDR3s associated with that allele. The upper chart represents alleles that have 100% identity to rhesus genomic reference sequence or previously published germline alleles. The lower chart includes only those alleles that have not been published previously and are defined as novel rhesus germline alleles.