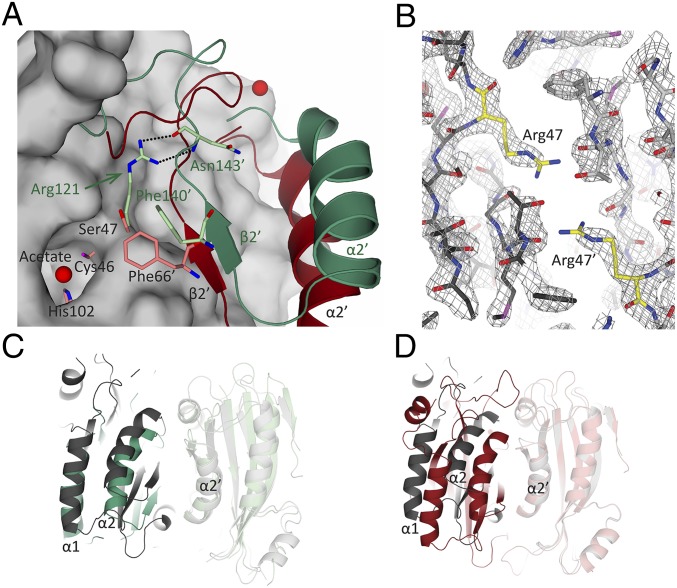
Fig. 5.
Comparison of the positioning of Arg121 and Ser47 in CrLCIB-ΔC, PtLCIB4, and PtLCIB4 S47R mutant. (A) The CrLCIB-ΔC dimer (green) and PtLCIB4 dimer (red) are aligned based on one subunit. The subunit used for alignment in the PtLCIB4 dimer is shown as a gray surface, excluding Zn, its binding residues (Cys46, His102, and Cys126), and Ser47. The residues from the other subunit in proximity to Arg121 (CrLCIB-ΔC) and Ser47 (PtLCIB4) are shown as sticks. The secondary structure elements are labeled in dark green (CrLCIB-ΔC) and black (PtLCIB). (B) Structure of the PtLCIB4 S47R dimer at the mutation site. The two subunits of the mutant dimer are depicted in dark gray and light gray, respectively. Arg47 from both subunits are colored yellow. The 2Fo-Fc electron density map (gray mesh) clearly shows the mutation of Ser47 to arginine. (C and D) The dimer of PtLCIB4 S47R (gray) compared with CrLCIB-ΔC (C, green) and PtLCIB4 (D, red) by alignment based on one subunit (shown in transparency).