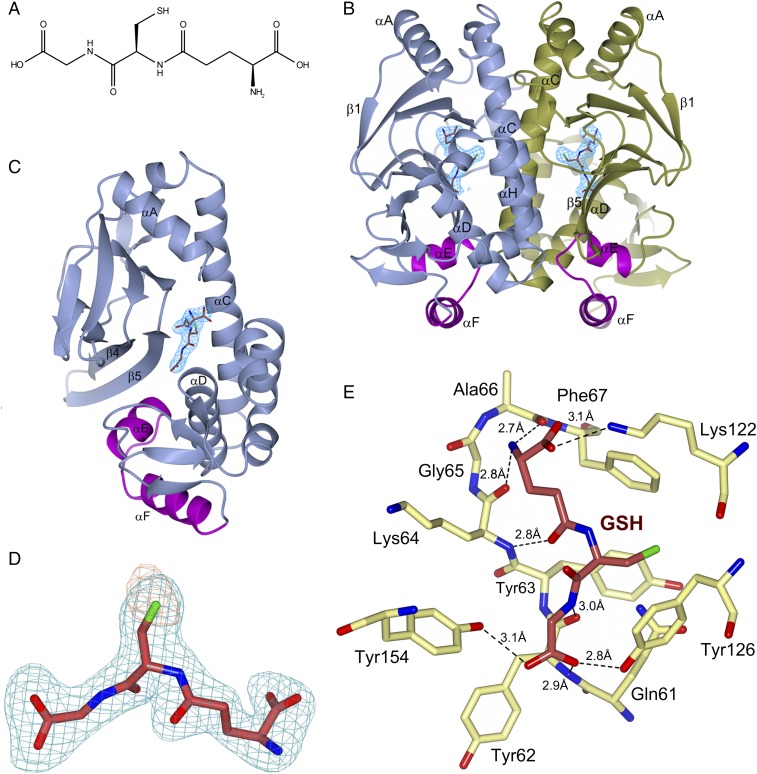
Fig. 1.
Structure of PrfAWT in complex with GSH. (A) Chemical structure of GSH. (B) Ribbon representation of the PrfAWT homodimer showing the binding sites of GSH at the tunnel site of each monomer. The monomers A and B are colored in blue and gold, respectively. The HTH motif is highlighted in magenta, and GSH is shown as sticks with carbon atoms colored crimson. The difference (2|Fo|-|Fc|) electron density map calculated from the refined PrfA-GSH complex and contoured at the RMSD value of the map is shown in blue covering GSH only. (C) Alternative orientation of B showing monomer A only. (D) Representative electron density covering the refined model of GSH in monomer A. An unbiased difference (|Fo|-|Fc|) electron density map contoured at three times the RMS value of the map is shown in blue. To reduce model bias, the GSH molecule was excluded from the coordinate file that was subjected to refinement before calculation of the electron density map. The anomalous log-likelihood-gradient (LLG) map, in orange, shows the position of the sulfur atom of GSH. (E) Key local structural features and amino acids forming direct hydrogen bonds (dashed lines) to GSH in monomer A. Gln123 and Trp224 are not shown for clarity. Additional structural features in the vicinity of the GSH-binding site are shown in Fig. S1.