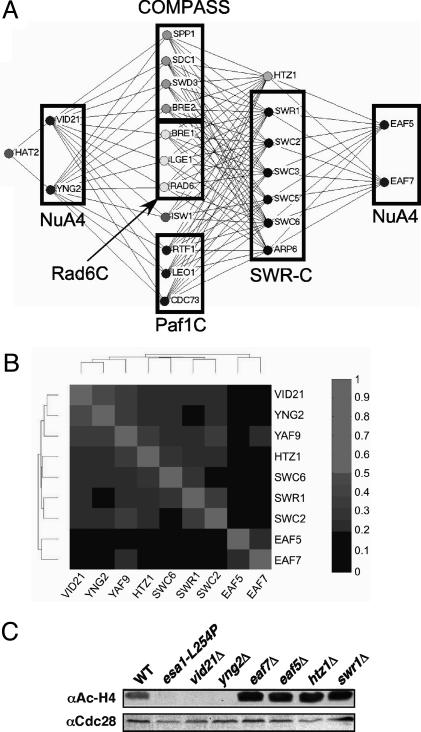
Fig. 2.
Genetic similarity of the SWR-C and a subset of NuA4. (A) Synthetic genetic interactions for the SWR-C and NuA4. SGA technology (19) was used to cross Natr strains harboring individual deletions of genes encoding Htz1 or subunits of the SWR-C or NuA4 with a transcription-targeted array of 384 KanR deletion strains to create sets of Natr Kanr haploid double mutants. Growth rates were assessed by automated image analysis of colony size. Lines connect genes with synthetic genetic interactions. The lengths of lines and proximity of boxes in this diagram and in Fig. 3A are unrelated to the strengths of the indicated synthetic genetic interactions. (B) Microarray analysis of gene expression was performed for the indicated deletion strains. Pearson correlation coefficients were then calculated for each pair of deletions, and the deletions were organized by 2D hierarchical clustering according to the similarities of their effects. (C) Immunoblots of whole-cell protein extracts from wild-type (WT, NJK28), esa1-L245P (LPY3500), vid21Δ (NJK1042), eaf7Δ (NJK1254), yng2Δ (NJK1482), eaf5Δ (NJK1259), htz1Δ (NJK1527), and swr1Δ (NJK1665) cells (see Table 3) grown to log phase in yeast extract/peptone/dextrose at 30°C and transferred to 37°C for 4 h. The membranes were probed with antihyperacetylated histone H4 (Penta) antibodies (α Ac-H4) and then reprobed with antibodies to Cdc28 to verify approximately equal protein loading.