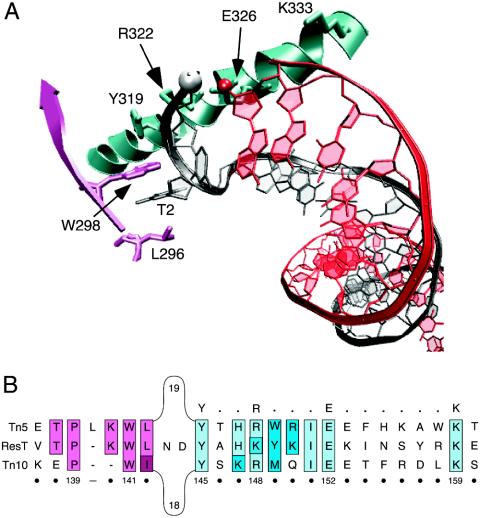
Fig. 1.
The hairpin binding motif of cut-and-paste transposases Tn5 and Tn10. (A) The hairpin binding domain bound to Tn5 hairpin DNA. The structural schematic is from the Tn5 crystal structure (Protein Data Bank ID code 1MUS) and was generated by using the vmd molecular graphics program (46). The 5′ phosphate and 3′ hydroxyl group that will take part in forming the interstrand bond are shown in the white and red spheres, respectively. The residue numbering indicated on the structure corresponds to that of the Tn5 transposase amino acid sequence. T2 is the flipped-out thymine base at position 2. (B) Amino acid sequence alignment of ResT (4, 13) with the cut-and-paste transposases of Tn5 and Tn10. Sequence analysis reveals that ResT contains a putative hairpin binding region similar to that found in the Tn5 (27, 28, 32) and Tn10 (31) transposases. The conserved Y-(2)-R-(3)-E-(6)-K signature found in the transposases of IS4 family members (33) is indicated in bold above the alignment. The residues that constitute the hydrophobic binding pocket are colored pink, and those contained in the Y-(2)-R-(3)-E-(6)-K signature are colored blue. Sequence identity is indicated by lighter shades; amino acids with similar properties are denoted by darker shades. The numbering below the alignment corresponds to positions in the ResT sequence.