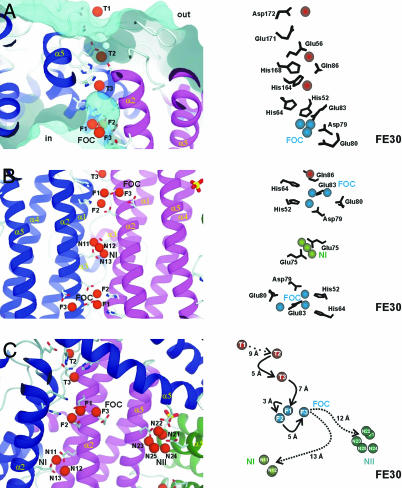
Fig. 2.
Overview on the iron-binding sites in H. salinarium ferritin based on the FE30 data set. (Left) The general environment of the iron atoms; (Right) detailed analysis of the residues involved and some distances following the iron entry to the final storage center are given. (A) Iron translocation over the DpsA protein shell from the outside (out) to the inner cavity (in). A surface representation of the outer and inner surface is shown in cyan. Side view of the iron translocation channel (from the FE30 dataset) with the initial iron-binding site T1 and the successive iron-binding sites T2 and T3, which occur in the iron translocation channel. The important side chains guiding the iron atoms are shown. (B) Two ferroxidase centers of symmetry-related protein molecules (colored in blue and red) are viewed along the twofold axis. The helices α1–α5, important residues, and the three iron-binding subsites F1–F3 of the FOC and nucleation center NI are depicted. Nucleation center NI, including the two symmetry-related iron atoms N11 and N13 and a third iron atom as well as the symmetry-related liganding residues Glu-72 and Glu-75, is shown. (C) View almost perpendicular to the threefold molecular axis. Overview of the ferroxidase and the two nucleation centers NI and NII. Possible routes for iron atom transfers from the FOC to NI and NII, respectively, are marked. Distances among iron centers and possible storage possibilities are indicated.