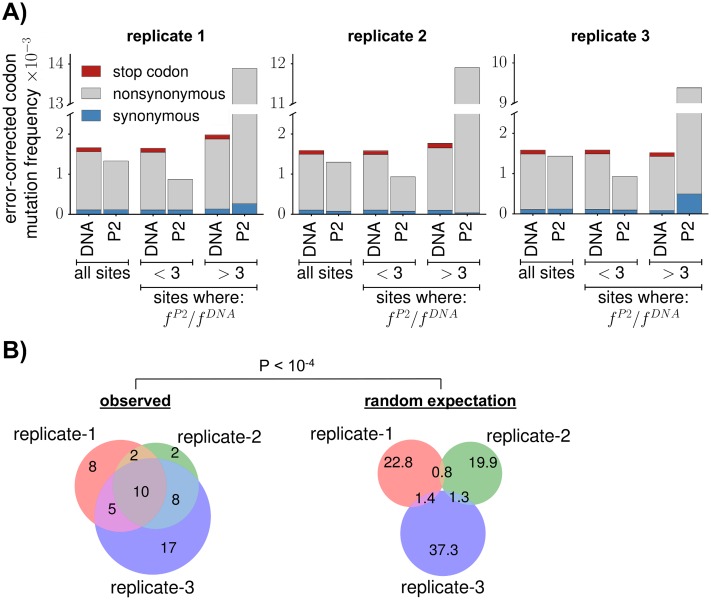
Fig 2. Selection purged mutations in most of env, but favored mutations at a few sites.
(A) For each replicate, we deep sequenced the initial plasmids (DNA) and the viruses after two rounds of passaging (P2). Bars show the per-codon mutation frequency averaged across sites after subtracting error rates determined from the wildtype controls (S2 Fig). When mutation frequencies are averaged across all sites, selection purged stop codons to <1% of their frequency in the initial DNA. Selection only slightly reduced the average frequency of nonsynonymous mutations; however, this average results from two distinct trends. For ≈4% of sites, the frequency of nonsynonymous mutations in the twice-passaged viruses (fP2) increased >3-fold relative to the frequency in the initial plasmid DNA (fDNA). For all other sites, the frequency of nonsynonymous mutations decreased substantially after selection. (B) The sites at which the error-corrected mutation frequency increased >3-fold are similar between replicates, indicating consistent selection for tissue-culture adaptation at a few positions. The left Venn diagram shows the overlap among replicates in the sites with a >3-fold increase. The right Venn diagram shows the expected overlap if the same number of sites per replicate are randomly drawn from Env’s primary sequence. This difference is statistically significant, with P < 10−4 when comparing the actual overlap among all three replicates to the random expectation. Another summary view of selection on env is provided by S4 and S5 Figs.