Abstract
High-throughput genotype screening is rapidly becoming a standard research tool in the post-genomic era. A major bottleneck currently exists, however, that limits the utility of this approach in the plant sciences. The rate-limiting step in current high-throughput pipelines is that tissue samples from living plants must be collected manually, one plant at a time. In this article I describe a novel method for harvesting tissue samples from living seedlings that eliminates this bottleneck. The method has been named Ice-Cap to reflect the fact that ice is used to capture the tissue samples. The planting of seeds, growth of seedlings, and harvesting of tissue are all performed in a 96-well format. I demonstrate the utility of this system by using tissue harvested by Ice-Cap to genotype a population of Arabidopsis seedlings that is segregating a previously characterized mutation. Because the harvesting of tissue is performed in a nondestructive manner, plants with the desired genotype can be transferred to soil and grown to maturity. I also show that Ice-Cap can be used to analyze genomic DNA from rice (Oryza sativa) seedlings. It is expected that this method will be applicable to high-throughput screening with many different plant species, making it a useful technology for performing marker assisted selection.
It is now routine for scientists to have the need to genotype thousands of individual progeny from populations of plants that are segregating one or more polymorphic loci. This type of analysis is performed by researchers investigating gene function using model systems, as well as by plant breeders pursuing marker assisted selection of crop plants (Koebner and Summers, 2003). In order to perform these genotyping experiments it is necessary to obtain tissue samples from individual plants in a manner that does not kill the plants. In addition, each tissue sample must be kept separate so that an accurate genotype for each individual can be determined. At present, there is no method available to perform high-throughput tissue sampling of individual plants. The current state-of-the-art consists of a technician with a paper punch collecting leaf cuttings one-at-a-time from individual plants. In this paper I describe a method that allows one to simultaneously harvest tissue samples from 96 individual plants. The method is highly parallel and is suitable for automation. Although this method was initially developed using Arabidopsis, I also demonstrate that the system can be used to analyze rice (Oryza sativa). It is expected that this method will be applicable to most plant species.
RESULTS
Overview of the Ice-Cap Method
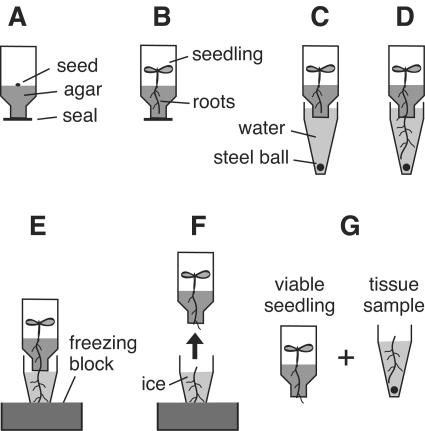
The method that I have developed for harvesting tissue from individual seedlings in 96-well format has been named Ice-Cap because ice is used to capture the tissue samples. A key component of this process is a 96-well plate with holes in the bottoms of the wells. Each well is shaped like a funnel as shown in Figure 1. The approximately 2.5-mm diameter holes in the bottom of this plate are initially sealed with a removable adhesive film, and agar growth substrate is deposited in the lower portion of the wells. A single seed is then deposited on the surface of the solidified agar in each well by using a novel seed loading device (Fig. 2). The seeds are allowed to germinate with the sealing film still covering the holes in the bottoms of the wells.
Figure 1.
Overview of the Ice-Cap procedure. A, Side-view of a single well from the 96-well seedling-growth plate. B, Following 3 d of growth under constant light the plants have germinated and roots have penetrated the surface of the agar. C, The upper 96-well plate is stacked on top of a second 96-well plate that is filled with water. D, The roots grow down into the water in the lower 96-well plate. E, The stacked plates are placed in a 96-well thermal block resting in a dry ice/ethanol bath. F, The water in the lower plate freezes in approximately 5 min, while the upper plate remains at room temperature. The upper plate is then separated from the lower plate. G, This process yields one 96-well plate containing root tissue samples and one 96-well plate containing viable seedlings.
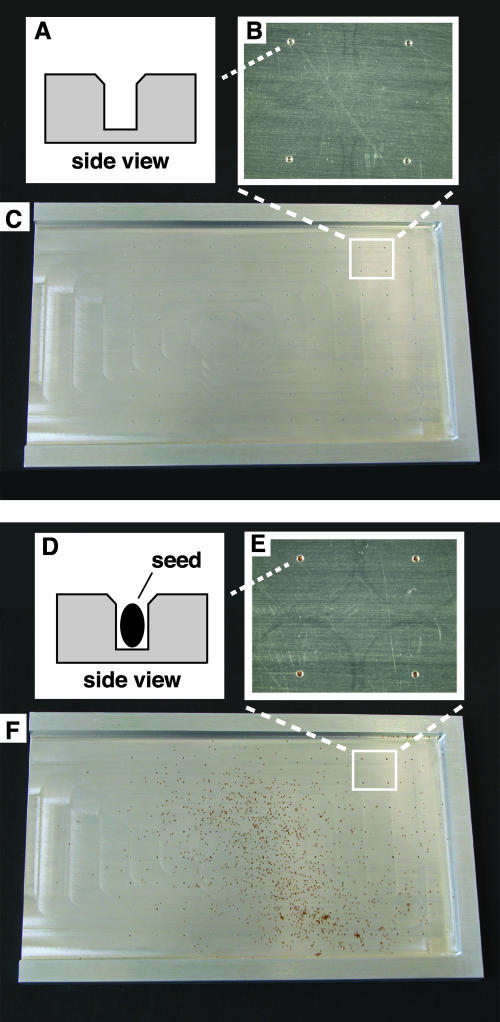
Figure 2.
The 96-well seed loading device. A, Side-view diagram of a single hole in the empty seed loader. B, Top view of four empty holes in the surface of the seed loader. C, Top view of the entire seed loader prior to loading of seeds. The three raised edges serve as guides that precisely position the 96-well plate on the loader. D, Side-view diagram of a single Arabidopsis seed occupying a hole in the seed loader. E, Top view of four holes in the surface of the seed loader. Each hole contains a single Arabidopsis seed. F, Top view of the entire seed loader during the seed loading process. Excess seeds can be seen littering the surface of the loader. The seed loader is 9.5 cm wide and 15 cm long.
Once the roots start to penetrate the agar substrate, the sealing film is removed from the bottom of the 96-well plate, exposing the agar. This plate containing the seedlings is then stacked on top of a second 96-well plate that contains water. The exposed agar surfaces of the upper plate are in contact with the water present in the wells of the lower plate. The plates are left stacked in this configuration approximately 2 weeks, during which time gravitropism causes the roots to grow down through the holes in the bottom of the upper 96-well plate and into the water-filled wells of the lower plate. The roots are allowed to grow until a significant amount of root tissue is present in the wells of the lower plate.
Tissue samples from each plant are then collected by transferring the stacked plates to a 96-well thermal block resting in a dry ice/ethanol bath. The wells of the lower plate are placed into the holes of the thermal block, which freezes the water in a manner of minutes. The upper 96-well plate, which is still stacked on top of the lower plate, remains at room temperature. Once the water in the lower plate has frozen solid, the upper plate containing the seedlings is peeled away from the lower plate. During this process the roots that are frozen in the lower plate break off and are trapped in the frozen water, while the seedlings and a portion of the root system remain in the upper 96-well plate. The seedlings in this upper plate are fully viable following this treatment. The final products of this process are two 96-well plates: one plate that contains 96 living seedlings and a second plate that contains 96 root tissue samples. The root tissue harvested using Ice-Cap can be used to perform high-throughput genotype analysis as described below.
96-Well Seed Loading Device
As described above, the Ice-Cap process involves growing seedlings directly in 96-well plates. In order to make the genotyping pipeline truly high-throughput it was necessary to develop a highly parallel method for depositing individual seeds into the wells of a 96-well plate. The seed loading device displayed in Figure 2 was fabricated in order to meet this need.
The seed loader is a rectangular metal plate with raised edges along three of the sides that allow a 96-well plate to be precisely aligned on the plate. The flat surface of the seed loader features an array of 96 holes that correspond to the locations of the wells in a 96-well plate. A variety of dimensions were tested for these holes in order to determine a size that was big enough for one Arabidopsis seed to fall into the hole, but not so big that two seeds could occupy it. The optimal hole diameter for Arabidopsis ecotype Columbia seeds was determined to be 400 microns, with a depth of 600 microns. An important feature of the holes in the seed loader is that they are chamfered, meaning that the diameter of the top portion of the hole is greater than the diameter of the lower portion, and tapered as shown in Figure 2A. It was found that this design was necessary to allow the oblong-shaped Arabidopsis seeds to easily fall into the holes. Another modification that further improved the unit's performance was the application of a coating of Paralyene to the surface of the seed loader. Paralyene is a specialized polymer that is deposited on the surface of metals through a process of gas-phase deposition to provide a low-friction coating. The Paralyene coating improves the ability of seeds to slide into and out of the holes in the seed loader.
To use the seed loader one begins with the unit resting on a flat surface with the holes facing up. Approximately 500 to 1,000 Arabidopsis seeds are placed on the loader, and one gently shakes the unit to move the mass of seeds around the surface of the plate. During this process seeds that happen to pass over the holes in the plate will fall into those holes. After shaking the unit for approximately 1 min, a tissue or small paintbrush is used to sweep excess seeds off the seed loader onto a sheet of paper. As shown in Figure 2D, the depth of the holes was chosen so that a single seed will be contained below the surface of the plate. If two seeds happen to fall into a hole, then the second seed will stick up above the level of the plate's surface. The process of brushing across the surface of the plate removes any excess seeds that protrude out of the holes.
Once the unit has been loaded with seeds, a 96-well plate containing agar growth media is placed upside down on top of the seed loader so that the open holes in the top of the growth plate are in contact with the seed loader. The stack is then carefully flipped over so that the seed loader is now sitting on top of the 96-well plate. A rubber mallet is then used to tap the seed loader, which causes the seeds to fall out of the holes in the seed loader and down onto the surface of the agar in the 96-well growth plate. Ideally the seed loader should distribute exactly 96 seeds into the 96 wells of the growth plate. In practice I have found that an average of 3 out of the 96 holes contains 2 seeds, corresponding to a single-seed plating efficiency of 97%.
Growth of Seedlings and Harvesting of Tissue
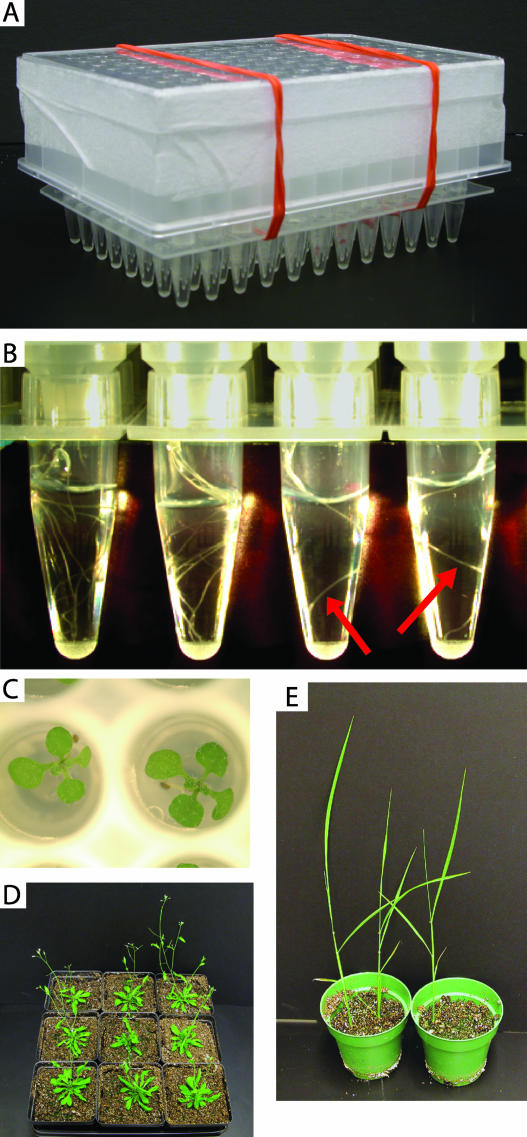
In order to test the performance of the Ice-Cap method, I used the seed loading device described above to plant Arabidopsis seeds into four 96-well plates. The seeds for this experiment were the progeny of a self-fertilized parent that was heterozygous for a T-DNA insertion in an Arabidopsis MAP kinase gene (MPK11, At1g01560; Alonso et al., 2003). The plants were grown under constant light at 21°C using the stacked 96-well plate arrangement described above and shown in Figure 3A. Following 14 d of growth under constant light, root tissue can be easily seen in the lower 96-well plates that contain water (Fig. 3B). A top view of the seedlings growing in the upper 96-well plate can be seen in Figure 3C. Fungal contamination was not observed in either the upper or lower 96-well plates, presumably because the seeds were surface sterilized prior to planting.
Figure 3.
Growth of Arabidopsis and rice plants. A, Picture of the Ice-Cap growth system with the stacked upper and lower 96-well plates visible. B, Close-up view of 4 wells of the lower 96-well plate into which roots have grown. Roots are visible in all 4 wells and are highlighted with red arrows in 2 of the wells. The nibs from the upper 96-well plate can be seen sticking down into the lower plate in this picture. C, Top view of 2-week-old Arabidopsis seedlings growing in the wells of the upper 96-well plate. D, Nine Arabidopsis seedlings were transferred from the 96-well growth apparatus into soil and grown under constant light. The pots are 9 cm wide. E, Three rice seedlings were transferred from the 96-well growth apparatus into soil and grown under constant light. The pots are 4 inches in diameter.
Ideally, all 96 wells of the growth plate would send roots down into the lower 96-well plate containing water. In this experiment the germination rate of the seeds was approximately 90%. Roots were found in the wells of the lower 96-well plate in all cases where a seedling had germinated and grown in the upper 96-well plate. The Ice-Cap method described above was then used to capture root tissue in the lower 96-well plates. The efficiency of root tissue harvest was 100%, meaning that all of the wells into which a root had grown yielded a root tissue sample. These results demonstrate that the growth system and tissue harvesting system are both very efficient and that the performance of the method is limited only by the germination rate of the seeds being used.
Following the harvest of root tissue, the 96-well plate containing the seedlings can be placed into a fresh 96-well plate that contains water. This plate should be set up in the same manner as the original root-collection plate. The purpose of this new plate is to keep the seedlings hydrated while you are performing the genotypic analysis. I have found that the plants remain viable for more than 1 week following Ice-Cap when stored in this manner at room temperature under constant light.
After tissue was harvested from the Arabidopsis seedlings using Ice-Cap, 9 plants were transferred from the 96-well growth plate into soil and grown under constant light. Seedlings were removed from the 96-well growth plate by placing a rubber hose over the opening in the bottom of the well containing the seedling of interest. The other end of the hose was connected to a pressurized air source, and the air pressure was carefully increased until the plug of agar containing the seedling slid out of the well onto the lab bench. The entire agar plug containing the seedling was transferred to soil, making no effort to remove agar from the root system. The transplanted seedlings were covered with a plastic dome to maintain high humidity for the first 2 d following transplant into soil. The transferred plants grew to full size in soil as shown in Figure 3D.
To determine if the Ice-Cap method could be used for species other than Arabidopsis, I tested the system using rice. Individual rice seeds were sown onto the agar surface of a 96-well growth plate. Following several days under constant light at 25°C, the roots of the rice seedlings easily grew down into the lower 96-well plate containing water. Root tissue was then harvested using the Ice-Cap method, with a 100% success rate. Three rice seedlings from which root tissue had been collected were then transferred to soil and grown under constant light. The rice plants grew well in soil following this treatment, as shown in Figure 3E.
DNA Preparation Using Ice-Cap Tissue Samples
A number of plant genomic DNA preparation protocols have been described in the literature (Aljanabi and Martinez, 1997; Lin et al., 2000; Xin et al., 2003). My goal was to identify a procedure that would yield usable DNA with a minimum of effort and expense. I tested a number of DNA extraction buffers and determined that the best results were obtained using a simple solution composed of Tris (pH 9) and EDTA. Because the root tissue samples are harvested in water, the first step of the DNA extraction process is to add concentrated Tris/EDTA to each well to bring the final concentration of the solution to 50 mm Tris, 5 mm EDTA. The 96-well plate is then sealed using a heat-sealing device and placed on a machine that rapidly shakes the plate, causing the metal ball present in each well to pulverize the root tissue. This step can be accomplished using either a modified paint shaker or a specialized tissue grinding machine. The metal balls used for tissue grinding are placed in the wells of the root harvesting plate during the initial set-up of the growth plate/root collection plate system. Following 5 min of shaking, the 96-well plate is placed in a centrifuge and spun for 10 min to pellet the pulverized root tissue. The supernatant containing genomic DNA is then removed from the plate using a pipette. This DNA solution is of sufficient purity to be directly added to a PCR.
I used quantitative, real-time PCR to determine the average yield of genomic DNA that was obtained using this protocol. For this experiment a pair of PCR primers was selected that amplify a 211-bp fragment of the Arabidopsis genome that is homozygous wild type in all of the plants used for this study. As a quantitative reference standard, I used a sample of Arabidopsis genomic DNA that had been purified using a high-yield DNA extraction protocol. The concentration of the DNA from this reference standard was determined using UV spectrophotometry. A series of PCR reactions were then set up that contained serial dilutions of this standard DNA, ranging from 85 ng down to 85 pg of genomic DNA per reaction. Similar PCR reactions were also set up in duplicate using DNA prepared as described above from eight different root tissue samples. I compared the threshold-cycle numbers determined via real-time PCR for each of the eight Ice-Cap DNA samples to the set of reference standards and determined that the average yield for the DNA preparation method was 400 ng of genomic DNA per sample, with a range of 100 to 700 ng. This quantity of genomic DNA is sufficient to perform hundreds of genotyping reactions.
To test the shelf-life of the genomic DNA prepared using this protocol I performed this same type of quantitative PCR experiment using aliquots of the root-tissue DNA samples that had been stored for 1 week at room temperature. No reduction in the quantity of PCR-detectable genomic DNA was observed following 1 week of storage at room temperature.
I also confirmed that the DNA preparation method was able to generate useful genomic DNA from rice. For this experiment a pair of PCR primers was chosen that amplify a 348-bp fragment of rice genomic DNA. PCR reactions were set up using DNA prepared using the Ice-Cap method, and PCR products of the expected size were generated. DNA sequencing confirmed that the amplified product was the expected rice genomic sequence.
Genotype Analysis
The Ice-Cap method was developed in order to eliminate a bottleneck that currently limits the efficiency of high-throughput genotyping in plants. It was therefore important to directly demonstrate that tissue harvested using Ice-Cap is suitable for use in genotyping. Towards this end I set up a genotyping pipeline that is based on the established method of melt-curve analysis, which was originally described as a technique for monitoring single-nucleotide polymorphisms (Akey et al., 2001; Lipsky et al., 2001). For my demonstration I am using melt-curve analysis to track a mutation caused by a T-DNA insertion. It should be noted, however, that any number of alternative genotyping methods could also be applied to tissue collected using Ice-Cap. Melt-curve analysis is presented here as one of many possible approaches.
In order to perform this genotyping experiment, four 96-well plates of Arabidopsis seedlings were processed using the Ice-Cap method. The parent plant that gave rise to this population of seedlings was heterozygous for a T-DNA insertion within the MPK11 gene (At1g01560). It is therefore expected that this population of seedlings will be segregating progeny that are homozygous wild type, heterozygous, and homozygous mutant at the MPK11 locus. Genomic DNA was prepared from each of the four plates of root tissue as described above, and these plates of genomic DNA were used to set up genotyping reactions.
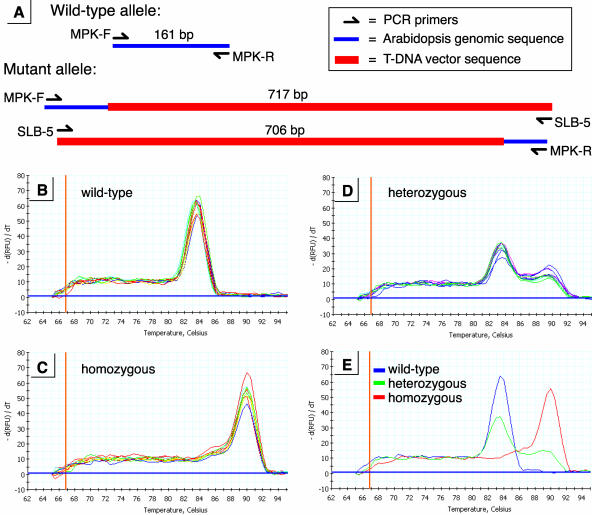
The melt-curve genotyping method I employed involved running a single PCR reaction using three PCR primers: MPK-F, MPK-R, and SLB-5 (Fig. 4A). Two of the PCR primers, MPK-F and MPK-R, hybridize to the MPK11 gene and amplify a 161-bp fragment of that locus in wild-type plants. In the T-DNA mutant allele these two primers are separated by greater than 10 kb of T-DNA, so that no PCR product is formed by this primer pair when only the mutant template is present in a reaction. The third primer present in the PCR reaction, SLB-5, hybridizes to a sequence found in the T-DNA vector's left border region. When the T-DNA mutant allele is present in the PCR reaction, then two different products are formed: SLB-5 + MPK-F yield a 717-bp product, while SLB-5 + MPK-R yield a 706-bp product (Fig. 4A).
Figure 4.
Genotype analysis. A, Maps of the PCR products produced by the wild-type and mutant alleles of the MPK11 locus. MPK-F, MPK-R, and SLB-5 are the three PCR primers used in the single-tube genotyping reaction. The total size of each PCR product is indicated in bp. The wild-type allele yields one PCR product, while the mutant allele yields two. B, Melt-curve analysis from nine representative plants that are homozygous for the wild-type allele of MPK11. C, Melt-curve analysis from nine representative plants that are homozygous for the mutant allele of MPK11. D, Melt-curve analysis from nine representative plants that are heterozygous at the MPK11 locus. E, Melt-curve peaks for a wild-type, a heterozygous, and a homozygous mutant plant are shown superimposed on a single graph. All of the melt-curve data display temperature on the x axis and change in fluorescence [−d(RFU)/dT] on the y axis. RFU, relative fluorescence units.
The locations of these PCR primers were chosen so that the PCR products produced by the mutant allele were much larger than the product formed by the wild-type allele. An important consequence of this size differential is that the resulting double-stranded PCR products will have significantly different melting temperatures. This situation makes it possible to use melt-curve analysis to determine the genotypes for each seedling (Akey et al., 2001; Lipsky et al., 2001). An attractive feature of melt-curve analysis is that it can be performed in the same 96-well plate that was used for the PCR amplification without the need for any further processing of the sample.
In order to measure the melt-curve profile for each PCR reaction, the reaction mixture included the DNA binding dye SYBR-Green. The amount of SYBR-Green fluorescence emitted by a sample is a direct measure of the quantity of double-stranded DNA present in that sample. The PCR reactions were run on a Bio-Rad iCYCLER iQ real-time PCR machine (Bio-Rad, Hercules, CA). Following PCR amplification, melt-curve analysis was performed. During this process the temperature of the samples is slowly raised from 65°C up to 98°C. SYBR-Green fluorescence is measured at 0.5°C intervals throughout this heating process. When the temperature of the wells reaches the Tm of one of the PCR products present in a sample, then there is a rapid drop in the SYBR-Green fluorescence emitted by that sample. The graphs shown in Figure 4 present the first derivative of the change in fluorescence displayed by each sample. This type of analysis produces a sharp peak corresponding to the Tm of the PCR product(s) present in a given sample.
The melt-curve profiles corresponding to wild-type, heterozygous, and homozygous samples are shown in Figure 4. A wild-type plant produces a single PCR product with a Tm of 83.5°C. Homozygous mutant plants produce a melt-curve profile with a single peak at 90°C. Although these reactions actually contain two different PCR products, the melt-curve profile resembles that of a single product because of the close similarity in size and sequence composition of the two products. As expected, heterozygous plants yield melt-curve profiles with two peaks, one corresponding to the wild-type allele, and one corresponding to the mutant allele. As shown in Figure 4E, these three genotypes can be easily distinguished using this simple, single-tube reaction.
The four 96-well plates of genomic DNA from the population of Arabidopsis plants segregating the mpk11 mutation were analyzed using the melt-curve method described above. A total of 347 genotypes were determined (Table I). Genotypes were not scored in wells where the seed did not germinate, or in those rare instances where two seeds were present in a single well. The distribution of homozygous, heterozygous, and wild-type genotypes was not significantly different from the expected Mendelian ratio. In order to confirm that the genotypes determined using the Ice-Cap method were accurate, I collected leaf tissue from 48 plants that had been genotyped using Ice-Cap. Genomic DNA was extracted from these leaf samples and PCR was performed using the same primer set described above. The genotypes determined using leaf tissue and those determined using Ice-Cap were in complete agreement.
Table I.
Genotype analysis using Ice-Cap
| Genotype | Plate 1 | Plate 2 | Plate 3 | Plate 4 | Sum | Percentage |
|---|---|---|---|---|---|---|
| Homozygous | 17 | 27 | 18 | 26 | 88 | 25 |
| Heterozygous | 49 | 36 | 46 | 40 | 171 | 49 |
| Wild type | 21 | 23 | 21 | 23 | 88 | 25 |
| Sum | 87 | 86 | 85 | 89 | 347 |
Four 96-well plates from a population of Arabidopsis plants segregating a T-DNA insertional mutation were processed using Ice-Cap and the genotype of each individual was determined as described in the text. The total number of genotypes successfully determined for each plate is indicated in the row labeled Sum. The column labeled Sum presents the combined data for all four 96-well plates. Percentage indicates the number of genotypes in a given category divided by 347, which is the total number of genotypes determined.
DISCUSSION
A valuable product of the genomic era will be an increasingly diverse collection of DNA markers that will allow plant breeders to precisely follow desired alleles through segregating populations (Peleman and van der Voort, 2003). As more and more molecular markers become available to the plant breeder, it will become increasingly valuable to be able to genotype large numbers of individual plants. For example, if a population of plants is segregating 2 alleles at each of 8 loci, then one would expect to find a plant that is homozygous at all 8 loci at a rate of 1 plant out of every 65,536 individuals. Given that many traits are controlled by quantitative trait loci, it is easy to imagine a plant breeder having the need to track many more than 8 loci in a segregating population. This situation highlights the need for an inexpensive, efficient system for genotyping large numbers of progeny. The Ice-Cap procedure that I describe in this paper should facilitate this type of analysis by allowing plant breeders to screen larger numbers of progeny at a lower cost than current techniques allow.
The Ice-Cap procedure should also assist scientists studying gene function using model systems such as Arabidopsis. Many classes of proteins in Arabidopsis are encoded by multi-gene families that are often comprised of more than 10 members. A popular method for analyzing gene function in Arabidopsis is to isolate knockout mutations in a given gene and observe the phenotype of a plant that is homozygous for that mutant allele. A common theme that is arising in the study of Arabidopsis genes is that functional redundancy within the members of a gene family often prevents a plant carrying a single homozygous knockout mutation from displaying a mutant phenotype. In order to overcome this functional redundancy, it is necessary to create plants that are homozygous for 2, 3, or more of the mutant loci (Liljegren et al., 2000; Pelaz et al., 2000; Bouche and Bouchez, 2001). If a population of plants is segregating mutant alleles at 5 different loci, then one would expect that the quintuple-homozygote would appear at a rate of 1 in every 1,024 plants. The Ice-Cap method would make searching for this rare plant a much more efficient process when compared to existing strategies.
Another application of Ice-Cap would be to screen large numbers of progeny in order to isolate individuals in which a rare recombination event has brought two closely-linked alleles into the cis-configuration. This type of screening would be useful in plant breeding as well as in gene-function studies using model systems. In either case, the identification of an individual that is homozygous at one of the loci and heterozygous at the second would indicate that recombination has brought the two mutant alleles together onto one of the chromosomes. When two desired loci are only a short distance apart on a chromosome, it can be necessary to screen thousands of progeny to find the rare recombinant.
Although Ice-Cap should be immediately useful for many plant species, the method cannot currently be used with plants that produce seeds that are too large to fit into standard 96-well plates. It should be possible, however, to design and construct large-format 96-well plates with wells that would be big enough to accommodate any size of seed. Although the perimeter of this type of plate would be larger than that of a standard 96-well plate, the holes in the bottoms of the wells could feed into angled channels that converge on the standard 96-well plate dimensions. In this manner a plate that is large enough to accommodate big seeds could still interface with a standard 96-well plate for root collection. This type of setup would allow the use of existing laboratory robotics and multi-channel pipetters when processing tissue samples collected from plants with large seeds.
The utility of the Ice-Cap method is not limited to genotyping. One could also use the tissue collected using Ice-Cap to perform high-throughput analysis of the metabolome, transcriptome, or proteome. Studies of this type could be used to perform forward-genetic screens on mutagenized populations in order to find rare individuals with a desired phenotype at the level of the metabolome, transcriptome, or proteome. In summary, the Ice-Cap method should provide plant scientists with a high-throughput platform for collecting tissue samples from large numbers of plants in order to perform any number of different types of downstream analyses.
MATERIALS AND METHODS
96-Well Seed Loading Device
The 96-well seed loading device was fabricated by V&P Scientific, San Diego (www.vp-scientific.com). Before using the seed loader, the Arabidopsis seeds were surface sterilized by soaking in 95% ethanol for 5 min and then allowed to air-dry overnight.
Plant Growth and Tissue Harvest
The upper 96-well plates in which the seeds were planted were purchased from Fisher Scientific and were Nunc brand, 1-mL filter plates without frit (Fisher catalog no. 278012). The holes in the bottoms of the plates were sealed using adhesive sealing film from Fisher Scientific (Fisher catalog no. 05–500–32). A total of 175 μL of autoclave-sterilized growth media (pH 5.8) composed of 0.5% (w/v) plant cell culture tested agar, 0.5× Murashige and Skoog basal salt mixture (Sigma-Aldrich, St. Louis, catalog no. M5524), and 2 mm MES (Sigma-Aldrich, catalog no. M-2933) was pipetted into each well of the plates with sealed bottoms. After the agar media had solidified, seeds were distributed into each well using the seed loader device. The tops of the 96-well plates were covered using clear plastic lids taken from Falcon Microtest flat bottom 96-well polystyrene plates (Falcon catalog no. 351172). The gap between the lid and the 96-well plate was then sealed using 3M Micropore surgical tape (Fisher catalog no. 19–027–761). The plates were incubated at 4°C in the dark for 4 d and then transferred to 21°C under constant fluorescent light.
After 3 d under constant light, the sealing film was removed from the bottoms of the 96-well plates. The 96-well plates containing the seedlings were then placed on top of 96-well plates that contained 275 μL of deionized water and a single metal ball in each well. The lower 96-well plates were Fisher thin-wall PCR plates with raised rims (Fisher catalog no. 0550127). The nibs from the upper 96-well plates were submerged in the water in the lower plates. The metal balls were either bismuth shot number 7 1/2 (Bismuth Cartridge, North Hollywood, CA) or 3/32-inch diameter stainless steel balls (Hartford Technologies, Rocky Hill, CT; catalog no. 034–006–1K). The stacked plates were held together using four rubber bands. The stacked plates were incubated under constant light for a total of 13 d, during which time the water level in the lower 96-well plates dropped below the level of the nibs from the upper plates. It is important that the humidity of the room in which the plants are grown is not too low. If the air is too dry it will cause the water in the root collection plate to dry out prematurely. If this problem presents itself then measures should be taken to raise the relative humidity of the air in the growth chamber or growth room.
To harvest root tissue from the plates, a 96-well thermal block was placed in a dry ice/ethanol bath and allowed to reach temperature equilibrium with the bath. The wells of the lower 96-well plate were then placed into the holes in the thermal block with the upper plate still held in place with the rubber bands. The stacked plates were left on the thermal block for 5 to 10 min, during which time the water in the lower plate froze solid. The stacked plates were then removed from the thermal block, and the rubber bands removed from the plates. The upper plate was then lifted off of the lower plate. The best approach to separating the two plates is to peel the upper plate away from the lower plate in the manner that the backing would be peeled off of a sheet of adhesive film. During this process the roots that are frozen in the lower plate break off at the level of the ice, leaving a sample of frozen root tissue in the lower plate and a viable seedling in the upper plate.
DNA Preparation
The 96-well plates containing the captured root tissue samples were left at room temperature until the ice in the wells had thawed. The average volume of water in the plates was 250 μL/well. Twenty-five microliters of 500 mm TRIS (pH 9) and 50 mm EDTA (pH 8) was added to each well. The plates were then sealed with Easy Peel heat sealing foil (catalog number AB–0745, ABgene, Surrey, UK) using a manual heat sealing machine (Thermo-Sealer, ABgene). The sealed plates containing root tissue, 275 μL of 50 mm TRIS and 5 mm EDTA, and a metal ball were loaded onto a 2000 Geno/Grinder machine (Spex/CertiPrep, Metuchen, NJ) and shaken for 5 min at a rate of 1,500 strokes/minute. After shaking the plates were spun in a centrifuge equipped with microplate carriers for 10 min at 3,500 rpm. The sealing foil was then peeled off the plates and 100 μL of supernatant was removed from each well by pipetting and transferred to a clean microplate. This supernatant contains the purified DNA extract and can be used directly in PCR reactions.
Genotyping Reactions
PCR reactions were performed using IQ SYBR Green Supermix (Bio-Rad). Each melt-curve genotyping reaction contained the following components: primer MPK-F at 0.1 μm, primer MPK-R at 0.25 μm, primer SLB-5 at 0.25 μm, 5 μL of IQ SYBR Green Supermix, and 1 μL of the genomic DNA solution isolated from root tissue (equal to 1–7 ng of genomic DNA, depending on the sample). The total reaction volume was 10 μL. The reactions were overlaid with 10 μL of mineral oil prior to running in the PCR machine. The following thermal cycling profile was run using an iCYCLER iQ real-time PCR machine (Bio-Rad): 95°C for 3 min 30 s; 45 cycles of 95°C for 10 s, 64°C for 1 min; 95°C for 1 min; 65°C for 1 min; melt curve analysis beginning at 65°C and increasing in 0.5°C increments for a total of 65 increments, holding each increment for 10 s. Primers used for the genotyping experiments were: MPK-F: TGCTCCTGAATGCAAACTGTGACCTCAAG; MPK-R: CCAAATGTCGATAGCTGCAGTGTATTCTG; and SLB-5: GGAAAACCCTGGCGTTACCCAACTTAATC. Primers used to measure the average yield of the genomic DNA preparation method were: MPK-16F: GATCATTTGGGATTCTTTCGAAACAGCTC and MPK-16R: CGTTGATAGGAATTCTCGACATTGGCATC. The primers used to amplify a fragment of the rice genome were RICE-F: CTGAAGATTCCATTAGGAGCGCAGTATTG and RICE-R: GATTGAGCGGCACGTTCTTTGAAATGTTC.
Acknowledgments
I thank Pete Jester and Sean Monson for technical assistance, Hye-Ran Lee for providing me with rice seeds, Patrick Cleveland for the Seed Loader device, and Bernadette Baker for coining the term Ice-Cap.
This work was supported by the U.S. Department of Agriculture (NRI, CSREES; USDA grant no. 2003–02593).
References
- Akey JM, Sosnoski D, Parra E, Dios S, Hiester K, Su B, Bonilla C, Jin L, Shriver MD (2001) Melting curve analysis of SNPs (McSNP): a gel-free and inexpensive approach for SNP genotyping. Biotechniques 30: 358–362, 364, 366–367 [DOI] [PubMed] [Google Scholar]
- Aljanabi SM, Martinez I (1997) Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res 25: 4692–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bouche N, Bouchez D (2001) Arabidopsis gene knockout: phenotypes wanted. Curr Opin Plant Biol 4: 111–117 [DOI] [PubMed] [Google Scholar]
- Koebner RM, Summers RW (2003) 21st century wheat breeding: plot selection or plate detection? Trends Biotechnol 21: 59–63 [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF (2000) SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404: 766–770 [DOI] [PubMed] [Google Scholar]
- Lin JJ, Fleming R, Kuo J, Matthews BF, Saunders JA (2000) Detection of plant genes using a rapid, nonorganic DNA purification method. Biotechniques 28: 346–350 [DOI] [PubMed] [Google Scholar]
- Lipsky RH, Mazzanti CM, Rudolph JG, Xu K, Vyas G, Bozak D, Radel MQ, Goldman D (2001) DNA melting analysis for detection of single nucleotide polymorphisms. Clin Chem 47: 635–644 [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Peleman JD, van der Voort JR (2003) Breeding by Design. Trends Plant Sci 8: 330–334 [DOI] [PubMed] [Google Scholar]
- Xin Z, Velten JP, Oliver MJ, Burke JJ (2003) High-throughput DNA extraction method suitable for PCR. Biotechniques 34: 820–824, 826 [DOI] [PubMed] [Google Scholar]