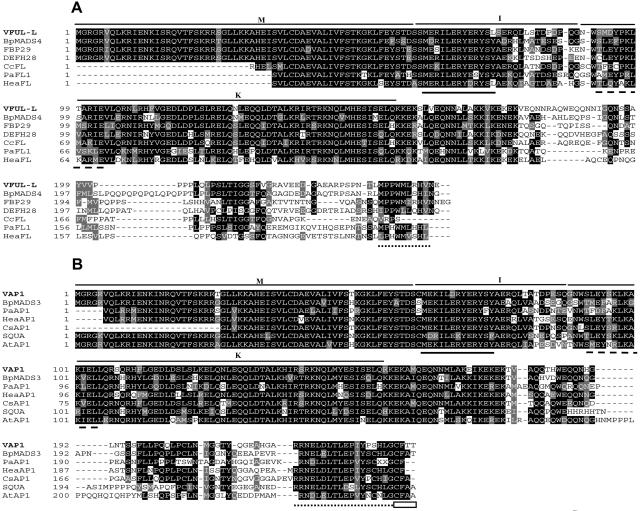
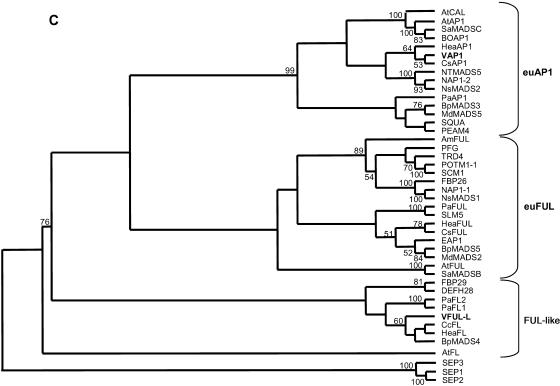
Figure 1.
Sequence analysis of VFUL-L, VAP1, and related SQUA-like proteins. A, VFUL-L protein and sequence comparison to other FUL-like proteins. B, VAP1 protein and sequence comparison to AP1/SQUA-like proteins. In both alignments, the MADS-box (M), I-region (I), and K-box (K) are indicated. In the I-region, the sequence highly conserved between both proteins is underlined with a continuous line. Dashed lines indicate distinctive sequences in the K-box. The highly divergent C-terminals show some conserved motives in each group (dotted line), mainly in the AP1 group that includes the prenylation motif (white box). C, Phylogenetic tree of the predicted SQUA-like proteins from different eudicots plant species (accession no. in parentheses): Antirrhinum SQUA, DEFH28, AmFUL (X63701, AY040247, AY306139); Arabidopsis AP1, CAL, FUL, AtFL (Z16421, L36925, AY072463, Q9LI72); Apple MdMADS2, MdMADS5 (U78948, AJ000759); Betula pendula BpMADS3, BpMADS4, BpMADS5 (X99653, X99654, X99655); Cauliflower BOAP1 (Z37968); Clarkia concinna CcFL (AY306143); Corylopsis sinensis CsAP1, CsFUL (AY306146, AY306147); Eucalyptus EAP1 (AF305076); Heuchera americana HeaAP1, HeaFL, HeaFUL (AY306148, AY306149, AY306150); Nicotiana sylvestris NsMADS2 (AF068726); Nicotiana tabacum NAP1-1, NAP1-2, NTMADS5 (AF009126, AF009127, AF068724); Petunia PFG, FBP26, FBP29 (AF176782, AY370517, AF335245); Pea PEAM4 (AF461740); Phytolacca americana PaAP1, PaFL1, PaFL2, PaFUL (AY306160, AY306161, AY306162, AY306163); Potato POTM1-1, SCM1 (U23757, AF002666); Silene latifolia SLM5 (X80492); Sinapis alba SaMADSC, SaMADSB (Q41276, Q41274); Tomato TRD4 (AY098732). Bootstrap support values are indicated when over 50. In order to root the SQUA subfamily, Arabidopsis SEP1 (M55551), SEP2 (M55552), and SEP3 (AF015552) were used as outgroup.