Figure 1.
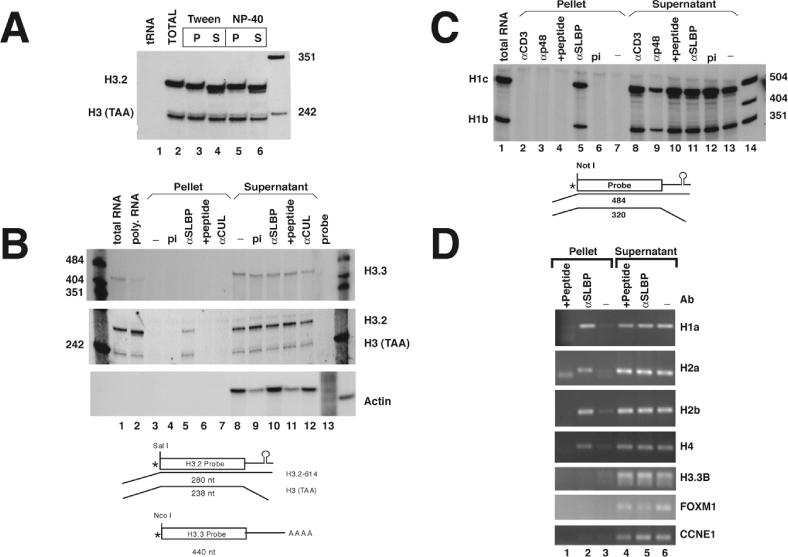
SLBP is bound to histone mRNAs in the cell. (A) Polyribosomes isolated from mouse myeloma cells were suspended in buffer containing 10 mM EDTA as described in Materials and Methods. α-SLBP was added in the presence of either 0.1% Tween-20 (lanes 3 and 4) or 0.1% NP-40 (lanes 5 and 6). RNA was prepared from the immunoprecipitates (lanes 3 and 5) and from the supernatant fraction (lanes 4 and 6) and analyzed by S1 nuclease mapping using the histone H3-614 gene as a probe. RNA prepared from polyribosomes was analyzed in lane 2 and yeast tRNA in lane 1. There is a small size differences (2–4 nt) observed between the total RNA (lane 2) and the immunoprecipitated mRNA compared with the RNA that did not immunoprecipitate. The final lane is pUC18 digested with HpaII. (B) Immunoprecipitations were performed from mouse myeloma polyribosomes with preimmune serum (pi) (lanes 4 and 9), α-SLBP (lanes 5 and 10), α-SLBP preincubated with 1 μg of antigenic peptide (lanes 6 and 11) or with an unrelated antibody, α-CUL (lanes 7 and 12). RNA was prepared from both supernatants and precipitates and analyzed for the polyadenylated H3.3 histone mRNA (upper panel) or the replication-dependent H3.2-614 mRNA (middle panel) by S1 nuclease mapping. The same RNA samples were analyzed for the mouse β-actin mRNA using a ribonuclease protection assay (lower panel). Analysis of total RNA or polyribomosal RNA are shown for H3.3 and H3.2 histone mRNAs but not for actin (lanes 1 and 2). A diagram of the S1 nuclease assay is shown below the figure. (C) Immunoprecipitations were performed with preimmune serum (pi) (lanes 6 and 12), α-SLBP (lanes 5 and 11), α-SLBP with antigenic peptide (lanes 4 and 10) or with the unrelated antibodies α-p48 and α-Cdk4 (lanes 2 and 3, and 8 and 9). RNA was prepared and analyzed by S1 nuclease mapping with the mouse H1c gene which maps both the H1c and H1b mRNAs (27). S1 nuclease mapping of total mRNA from the same cells is shown (lane 1). Lanes 7 and 13 shows polyribosomes incubated in buffer and then treated with protein A beads. Lane 14 is marker pUC18 digested with HpaII. A diagram of the S1 nuclease assay is shown below the figure. (D) Immunoprecipitations were performed from polyribosomes isolated from HeLa cells with either no antibody (lanes 3 and 6), α-SLBP (lanes 2 and 5) or α-SLPB with antigenic peptide (lanes 1 and 4). RNA was prepared and analyzed by RT–PCR with primers specific to H1a (HIST1H1A), H2a (HIST1H2AB), H2b (HIST1H2BG) and H4 (HIST1H4B). The same RNA samples were assayed for the presence of the polyadenylated histone H3.3 mRNA (H3F3B) and two additional cell-cycle-regulated mRNAs, FOXM1 and CCNE1 (62). PCR products were detected by ultraviolet (UV) illumination of ethidium bromide stained 2% agarose gels.
