Figure 2.
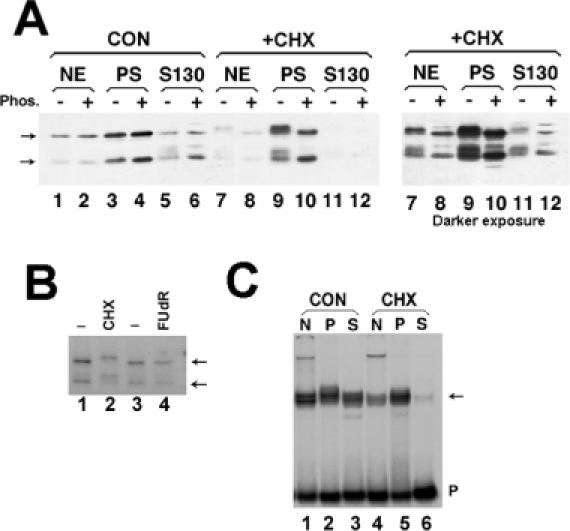
Effect of protein synthesis inhibitors on SLBP modification and localization (A) Mouse myeloma cells were grown normally (lanes 1–6) or treated with 0.1 mM CHX for 1 h (lanes 7–12). Both cultures were fractionated in parallel into nuclear extract (lanes 1, 2, 7, 8), polyribosomes (lanes 3, 4, 9, 10) or post-polyribosomal supernatant (S130, lanes 5, 6, 11, 12). The right hand panel is a longer exposure of the CHX fractionation (lanes 7–12). Extracts from equivalent amounts of cells were analyzed by gel electrophoresis and the SLBP detected by western blotting. There was some proteolysis of SLBP resulting in removal of ∼10 kDa from the N-terminus (18). Aliquots of the fractions in even-numbered lane were treated with protein phosphatase prior to electrophoresis. The position of unmodified SLBP and the proteolytic product are indicated by the arrows. (B) Mouse myeloma cells were labeled with H3[32PO4] and left untreated (lanes 1 and 3), treated with CHX for 2 h (lane 2) or treated with the DNA synthesis inhibitor FudR for 1 h (lane 4). Total cell lysates were prepared and the SLBP immunoprecipitated and resolved by SDS-PAGE. Phosphorylated SLBP was detected by autoradiography. The position of unmodified SLBP and the proteolytic product are indicated by the arrows. (C) The nuclear, polyribosomal and S130 extracts from equivalent amounts of mouse myeloma cells [same extracts as in panel (A)] were assayed by mobility shift assay using a radiolabeled stem–loop RNA as a probe (40). The stem–loop/RNA complex [(C) arrow] was resolved by native polyacrylamide gel electrophoresis.
