Figure 6.
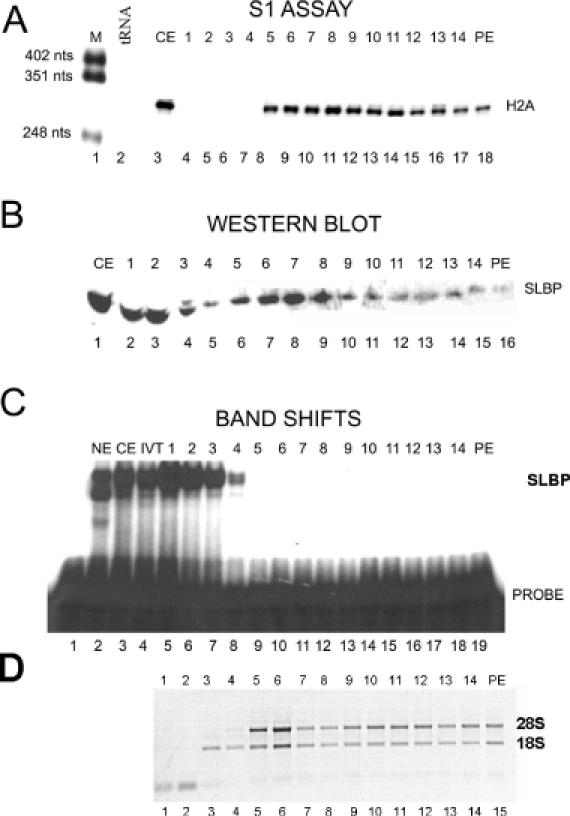
SLBP is associated with histone mRNA on ribosomes from CHX-treated cells. Cytoplasmic lysate prepared from HeLa cells treated with CHX for 45 min were fractionated by sucrose gradient centrifugation. The fraction numbers from the sucrose gradients are indicated above each lane; PE is the sucrose gradient pellet. Fractions 6 and 7 contain the monoribosome peak as determined by ethidium bromide staining to total RNA. (A) Total RNA isolated from each fraction was analyzed for histone H2a mRNA by S1 nuclease mapping. Lane 1, marker (pUC18 digested with HpaII); lane 2, 10 μg of yeast tRNA; lane 3, 5 μg of RNA from the cytoplasmic extract; lanes 4–18, sucrose gradient fractions. (B) Protein from each fraction was analyzed by western blotting for SLBP. Lane 1, 10 μg of cytoplasmic extract; lanes 2–16, sucrose gradient fractions; PE, sucrose gradient pellet. (C) Each fraction was analyzed for RNA binding activity by a mobility shift assay using the radiolabeled stem–loop RNA probe. Lane 1, probe; lane 2, nuclear extract; lane 3, cytoplasmic extract; lane 4, in vitro translated SLBP; lanes 5–19, sucrose gradient fractions. (D) An aliquot of each fraction was analyzed by agarose gel electrophoresis and the RNAs detected by ethidium bromide staining. Lanes 1–15, sucrose gradient fractions 1–14 and the pellet (PE).
