Figure 1.
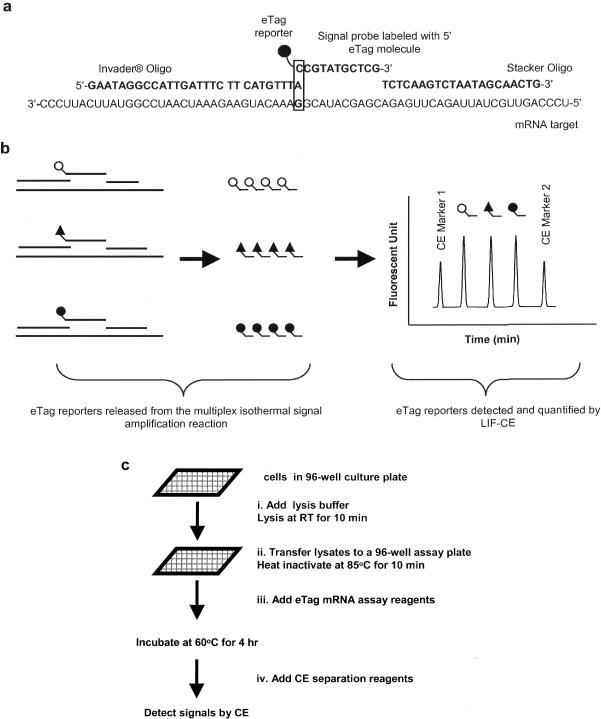
(a) Schematic representation of eTag multiplex mRNA assay coupling the eTag reporter chemistry and the Invader® mRNA reaction. A probe set is shown (from left to right) including Invader® oligonucleotide, signal probe coded with distinct eTag molecule and stacker oligonucleotide specifically designed to anneal to its respective RNA target. The 3′ terminus of Invader® oligonucleotide invades one base (non-complementary to template) into DNA–RNA duplex between signal probe and target, forming an overlapped DNA–RNA triplex structure shown in box. The Cleavase® enzyme, which possesses 5′ nuclease activity, recognizes and cleaves this specific structure, generating an eTag reporter (eTag molecule with 5′ terminal nucleotide). Subsequently, signal probes, in large excess, rapidly undergo association with the mRNA target replacing cleaved signal probes. Multiple signal probes are cleaved per RNA target, resulting in target-specific accumulation of eTag reporters. (b) Multiplex assay is achieved by allocating unique eTag molecules to specific probe sets. Multiple cleaved eTag reporters are separated by CE. (c) eTag multiplex mRNA assay procedure. Assay samples could be total RNA or crude lysate. There are four liquid addition steps starting from sample plate to CE analysis: (i) lysis buffer addition (this step is omitted if the sample is RNA), (ii) transfer the lysate from culture plate to a 96-well assay plate, (iii) reaction mixture (containing probes and enzyme) in addition to the assay plate and (iv) CE marker addition.
