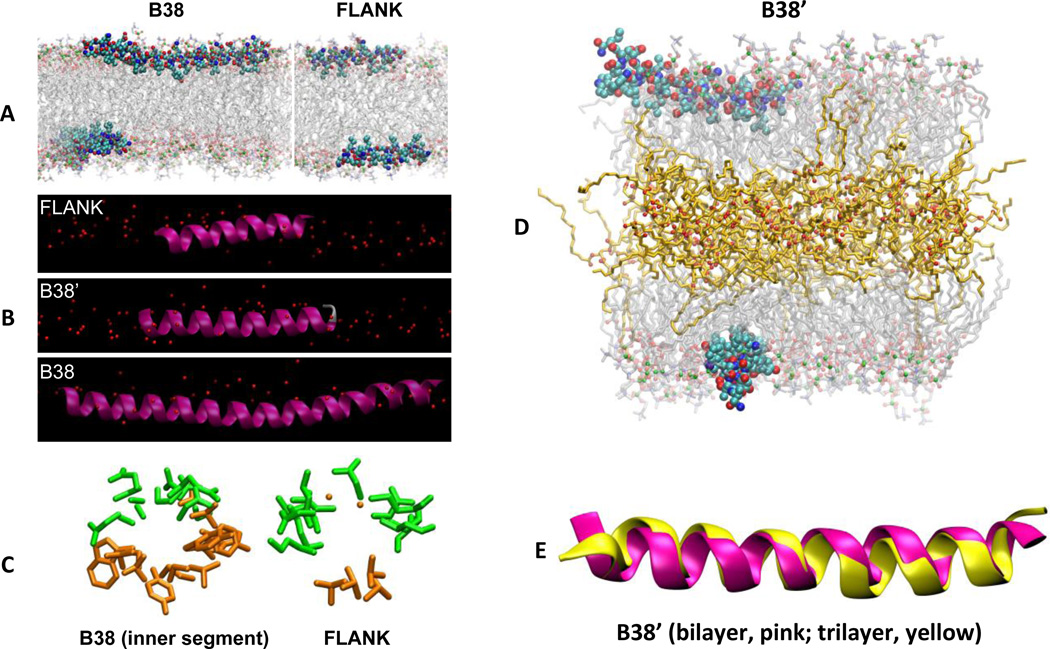
Figure 6. All-atom MD simulations of FLANK and B38 in a POPC bilayer.
Simulation snapshots of the peptides in their average orientations in three different representations. (A) B38 (left) and FLANK (right) in CPK representation with carbons light blue, oxygens red, and nitrogens blue. Acyl chains and head groups of lipids are represented by transparent lines and balls, respectively, with phosphorus atoms green. Water molecules and hydrogens are omitted for clarity. (B) FLANK, B38’, and B38 in the lipid surface (red spheres denote phosphorus). (C) “Helical-wheel” representations of peptides looking along the helical axes of B38 and FLANK. Only side chains (excluding hydrogen atoms) are shown for clarity. Hydrophilic and hydrophobic residues are shown in green and orange, respectively. These helical wheels, which are obtained using MD simulations, confirm those of Fig. 3, which are obtained using the Position of Proteins in Membranes server. (D) Simulation snapshot of B38’ interacting with a trilayer at 120 ns. Triglyceride acyl chains and oxygens are represented by orange lines and red balls, respectively. (E) Superimposed structures of B38’ in bilayer (pink) and trilayer (yellow). The structures correspond to last configurations of bilayer (520 ns) and trilayer (120 ns) systems.