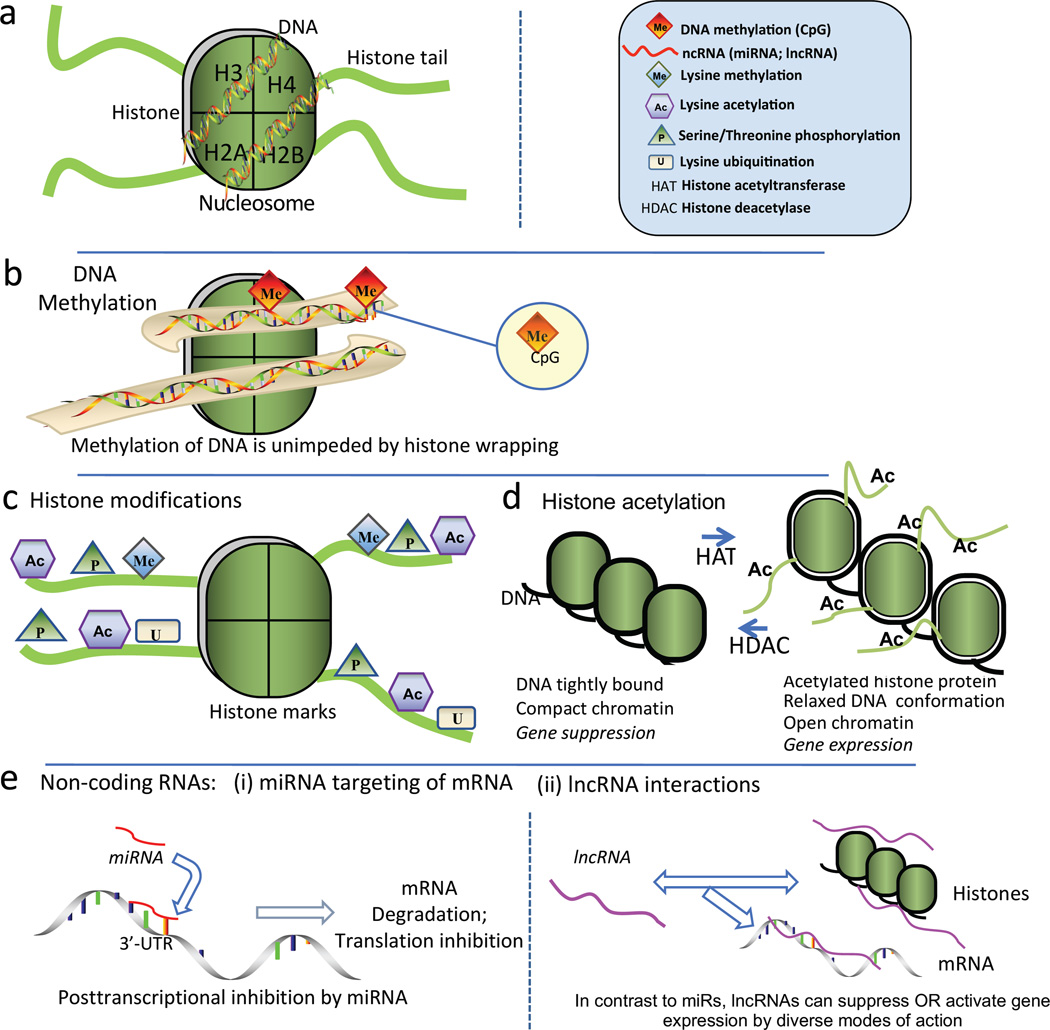
Figure 4.
a. Nucleosome structural elements. DNA is wrapped around each histone octomer protein that has basic amino acid-rich tail domains. Each core contains two copies of the histones H2A, H2B, H3 and H4, the amino terminal domains of which extend from the core as basic amino acid-rich ‘tails’ that can be modified, principally by acetylation and methylation (see Figure 4c) b. Proximal promoter DNA methylation at CpG islands results in gene silencing by transcriptional repression (cf. Fig 4b). c. Posttranslational ‘marks’ on N-terminal histone tails constitute a histone code; they include lysine acetylation and methylation, serine phosphorylation and lysine ubiquitination and sumoylation that remodel chromatin by changing histone protein-DNA association to influence transcriptional regulation. d. Chromatin remodeling by HATs and HDACs. Histone acetylation relaxes tightly wound compact DNA to enable access of transcriptional regulators. HDACs deacetylate to restore compact chromatin and prevent transcription. e. Posttranscriptional RNA-based mechanisms include short (microRNAs) and long noncoding RNAs (lncRNAs). e[i].miRNAs target the 3’-untranslated region of mRNA promoting transcript degradation and also translation destabilization. e[ii]. lncRNA facilitate changes of histone methylation through complex mechanisms that can result in enhanced or repressed gene expression depending upon the histone mark. Modified from Jiang et al (2015b).