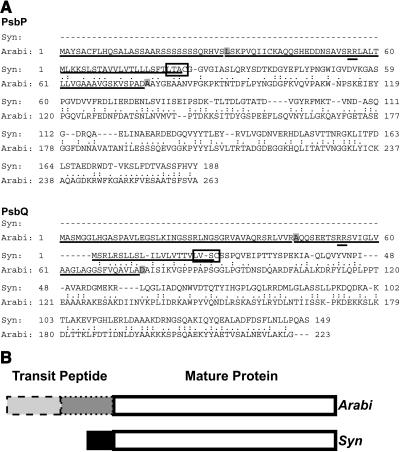
Figure 1.
Sequence Comparison of PsbP and PsbQ Proteins from Synechocystis 6803 and Arabidopsis.
(A) Alignments of the predicted amino acid sequences of PsbP and PsbQ proteins from Synechocystis 6803 (Syn) and Arabidopsis (Arabi). Similar residues are indicated by (.) and identical residues by (:). Targeting sequences are underlined. Synechocystis 6803 lipoprotein recognition sites are boxed, with cleavage before the Cys residue. Arabidopsis chloroplast transit peptide cleavage sites (as predicted by ChloroP) are in the first gray box, and thylakoid transit peptide cleavage sites (as predicted by SignalP and determined by N-terminal sequencing by Gomez et al., 2003) are in the second gray box in each sequence. The precursor forms of PsbP and PsbQ proteins in Synechocystis 6803 are 18 and 17.6% identical to their respective homologs in Arabidopsis in this global alignment, respectively.
(B) Diagram depicting sequence alignments of Synechocystis 6803 and Arabidopsis PsbP and PsbQ proteins. Both PsbP and PsbQ have bipartite transit peptides in Arabidopsis, one part for chloroplast envelope translocation (dashed outline) and one for thylakoid lumen localization (dotted outline). These sequences share very little similarity with the much shorter signal peptides on the Synechocystis 6803 proteins (black box). The white boxes represent the mature protein sequences for Arabidopsis and Synechocystis 6803 that are 27.3 and 32.9% identical for PsbP and PsbQ, respectively.