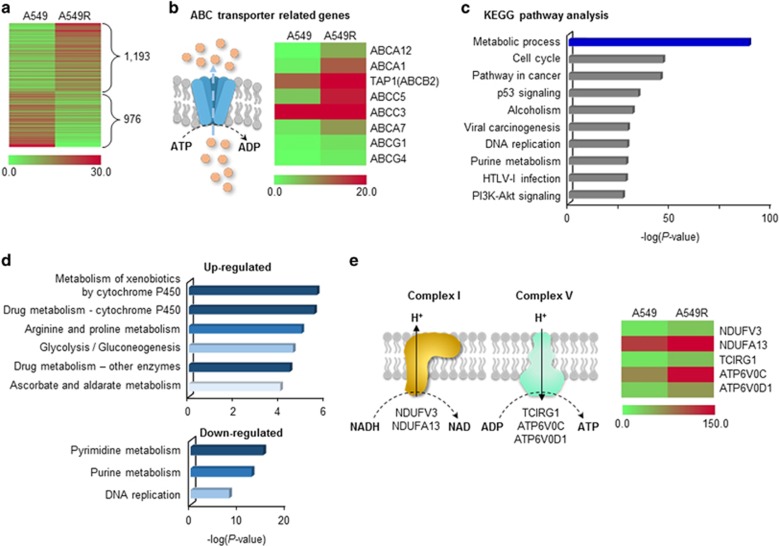
Figure 5.
Global changes in the expression of metabolism genes in cisplatin-resistant cells. (a) Differentially expressed genes (DEGs) between parental and cisplatin-resistant cells (A549 and A549R cells) were analyzed by RNA sequencing. Genes whose expression changed more than twofold are shown. (b) Drug-efflux systems were activated in A549R cells. Various ABC transporters were upregulated in A549R cells. (c) Metabolic process was the dominant pathway altered in A549R cells according to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. The top 10 pathways are presented; full analysis results are shown in Supplementary Table 2. (d) Drug detoxification and DNA synthesis-related metabolism were up- and downregulated, respectively, in A549R cells. Up- and downregulated DEGs in the metabolic process category were subjected to KEGG pathway analysis. Pathway maps with a P-value <1 × 10−4 are shown. (e) Genes related to OxPhos-complexes I and V were upregulated in A549R cells. All DEGs related to the OxPhos complexes whose expression changed more than twofold are shown.