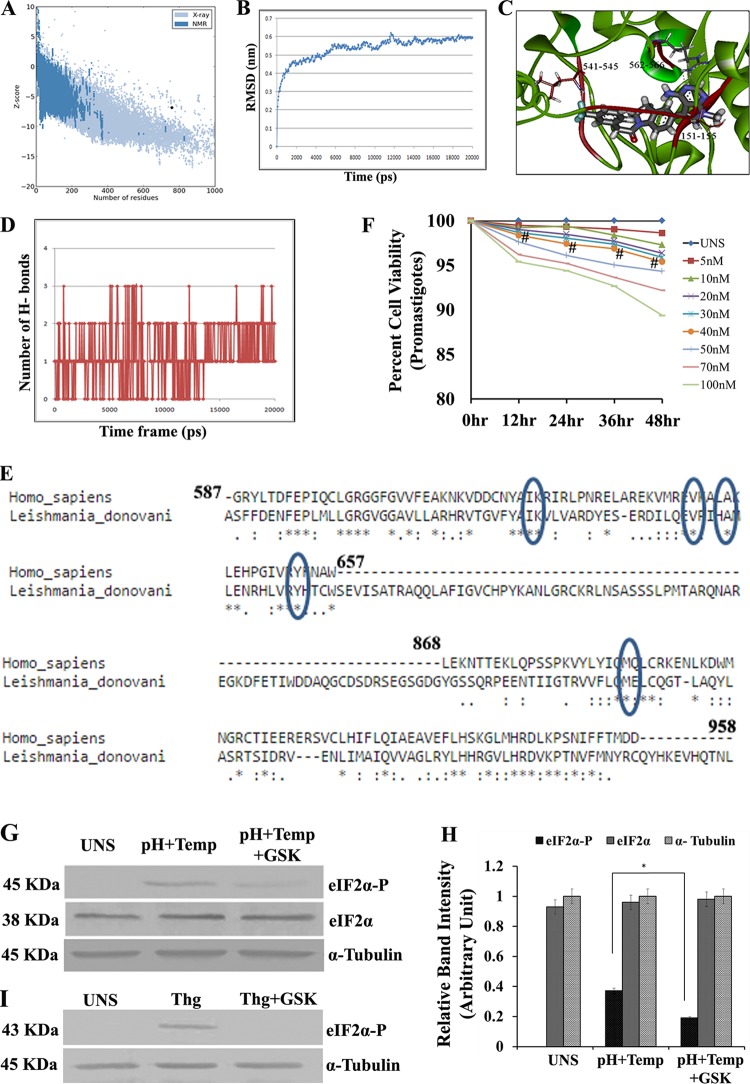
FIG 4.
Docking study of GSK2606414 as a PERK inhibitor. (A) Pro SA-web server. Plot validation of the modeled protein with the ProSA-web server showed a Z score of −6.84. The ERRAT plot showed that 86.46% of residues were below the error value cutoff limits. (B) An MD simulation was conducted for the modeled system in explicit solvent by using GROMACS 4.0.3. The RMSD in the C-α backbone atom in the L. donovani eIF2α kinase protein (LdeK) for up to 4 ns after the 20-ps model attains its maximum stability is shown. lsq, least squares. (C) Study of docking of GSK260414 (inhibitor of PERK enzymes) with the L. donovani eIF2α kinase protein. The result obtained from DS2.5 and Glide v9.10 suggest that the interaction between Leu151-Ser155, Arg541-Asp545, and Arg562-Arg566 may play a key role in the inhibition of L. donovani eIF2α kinase (LdeK). (D) Profile of H-bonding between GSK2606414 and the PERK kinase domain (LdeK) gene. (E) Comparison of L. donovani PERK and mammalian PERK (human) by protein sequence alignment. (F) Dose-response curve for different concentrations of GSK2606414. At concentrations of up to 40 nM, the percent viability of untreated promastigotes was above 95% compared to untreated parasites (UNS) (# denotes nonsignificant data sets compared with untreated ones). (G) Western blotting to determine the phosphorylation status of LdeIF2α under unstressed conditions, pH and temperature stress (pH+Temp), and pH and temperature stress after GSK2606414 (30 nM) treatment (pH+Temp+GSK). α-Tubulin was used as a control. (H) Densitometric analysis of band intensities. (I) Western blotting to determine the phosphorylation status of mammalian eIF2α (mouse PECs) under unstressed conditions, with thapsigargin treatment (1 μM), and in PECs treated with thapsigargin along with GSK2606414 (30 nM) (Thg+GSK) to show the effect of GSK2606414 on mammalian PERK. α-Tubulin was used as a loading control. Data displayed here represent results from one of three separate experiments.