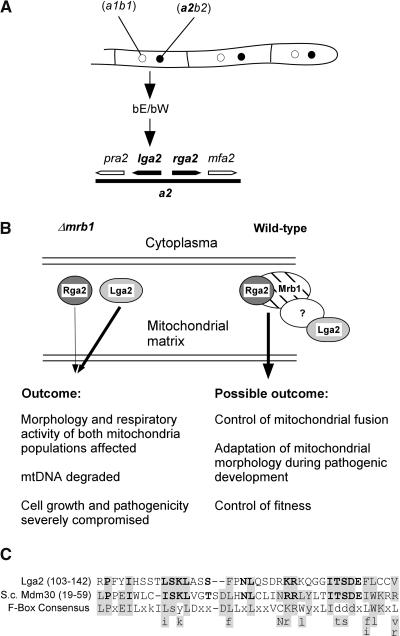
Figure 9.
Model for the Interplay among Mrb1, Rga2, and Lga2.
(A) Arrangement of the lga2 and rga2 genes on the a2 mating-type locus and direct control of lga2 expression by an active bE/bW complex formed in the dikaryon.
(B) In Δmrb1 mutants, Lga2 and Rga2 interfere with mitochondrial fusion in an uncontrolled manner and thus affect cellular fitness and fungal virulence. The extent of negative contribution relies on the observed genetic relationship and is reflected by the different thickness of the arrows. Although the presence of Lga2 alone is already sufficient to compromise pathogenicity, Rga2 can only enhance this effect in an Lga2-dependent manner. In wild-type cells, Mrb1 directly controls Rga2, whereas Lga2 is indirectly controlled, possibly by complex formation with a yet unidentified protein. The complex is presumed to control mitochondrial fusion and to be needed for adaptation of mitochondrial morphology and function in response to special demands during the host interaction. It may thus provide for control of fungal fitness in planta.
(C) The putative F-Box motif of Lga2 is aligned with the respective sequence of Mdm30 (S. cerevisiae) and an F-box consensus sequence (Patton et al., 1998). Residues that match F-box sequences are shaded. Identical or highly similar amino acids between Lga2 and Mdm30 are in boldface. Gaps have been introduced at a variable position of the F-box consensus sequence to increase the alignment. Small letters refer to amino acids that are not predominant in a position. Letters beneath the consensus refer to amino acids that fit the Lga2 or Mdm30 motifs and are found in at least two other F-boxes (Bai et al., 1996; Patton et al., 1998).