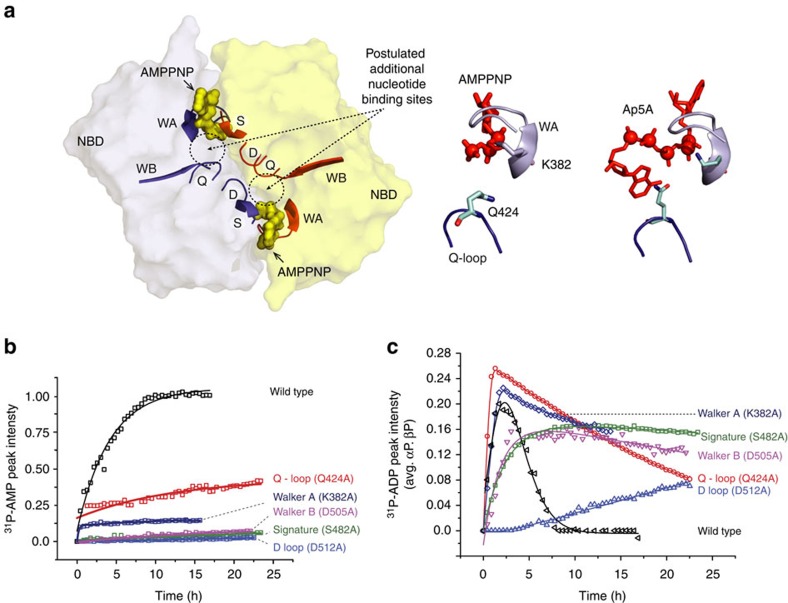
Figure 5. Mutational analysis of the coupled ATPase-AK activity of MsbA.
(a) Visualization of the conserved NBD sequence motifs in the MsbA/AMP.PNP crystal structure (pdb: 3B60). The canonical binding sites and a second postulated binding sites are highlighted. Key residue K382 in Walker A is in close proximity to the nucleotide phosphate groups (top middle). In pfSMCNBD, the Q-loop glutamine was found coordinating binding of Ap5A indicative of a second binding site (top right, pdb: 3KTA). All single-point mutations are summarized in Table 1. (b) Progress curves of the 31P-AMP peak intensities demonstrate much reduced generation of AMP. The initial data points of Q424A are missing due to the experimental dead time. (c) Progress curves of the 31P-ADP peak intensities are sensitive to both the ATPase (positive slope) and kinase (negative slope) reactions. The mutation Q424A causes accelerated ADP generation (ATPase) but slower consumption (kinase) with respect to wild-type MsbA. The ADP consumption is even slower in case of K382A. The S482A and D505A mutations slow down both ATPase and kinase activity. The D512A mutation causes the slowest ADP generation, whereas no consumption of ADP could be seen on the time scale of the experiment. Experiments were done in triplicate with protein samples from different expressions each time.