Figure 2.
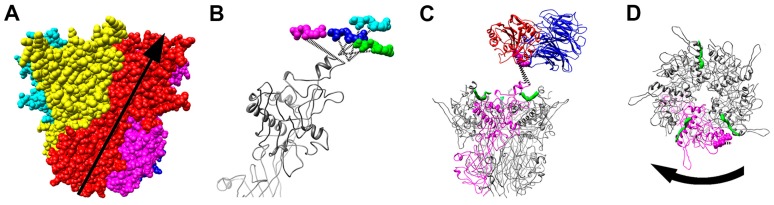
Natural twist of the penton base and possible untwisting by integrin. (A) Space-filling representation of the HAdV-C2 penton base pentamer (PDB-ID 1X9T) with each subunit in a different color. The natural twist of the penton base from the bottom of the pentamer to the solvent accessible surface in the virion is represented by an arrow; (B) Side view of four superimposed penton base monomers with four copies of the RGD residues (magenta, cyan, blue, and green) as modeled in the HAdV-A12/αvβ5-integrin cryoEM density. The dashed lines represent the extension of the flexible RGD loops. Note that the magenta copy of the RGD residues extends the RGD loop counter to the natural twist of the penton base subunit; (C) Side view of the penton base pentamer with one subunit in magenta, fiber N-terminal peptides in green, the magenta copy of the RGD residues as shown in panel B, and docked RGD-binding integrin domains (blue and red); (D) Top view of the penton base pentamer with a curved arrow indicating the direction of the possible untwisting precipitated by interactions with integrin. Reprinted with permission from [47]. Copyright © American Society for Microbiology, Journal of Virology, 83, 2009, 11491–11051, doi:10.1128/JVI.01214-09.