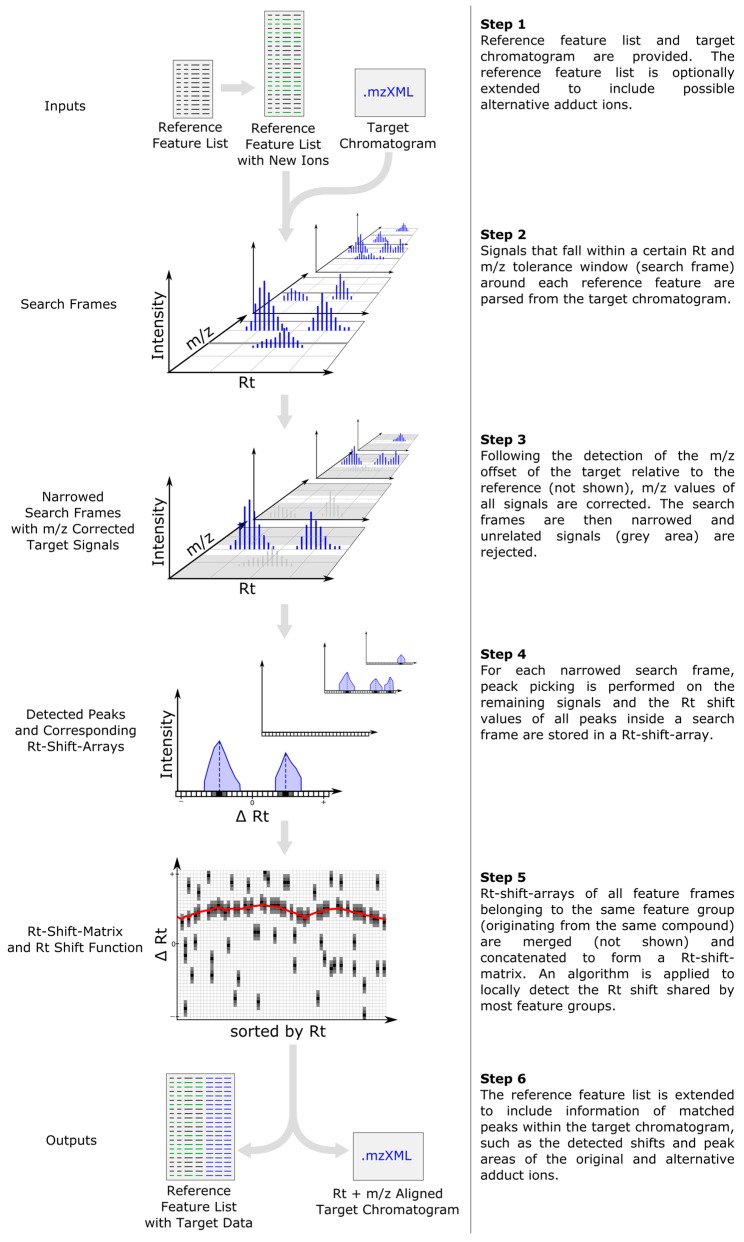
Figure 1.
Workflow depicting the main data processing steps of MetMatch for a single target chromatogram. If more than one target chromatogram is going to be processed, each of those is successively carried through the steps 2–5 and a single data table containing the (extended) reference list as well as the matched features of every target chromatogram is generated. After processing of the target chromatograms, all new information (chromatographic peak area, corrected m/z value and retention time, calculated m/z and Rt shift) for each reference feature and processed target chromatogram are saved as an extended data matrix. Moreover, the target chromatograms can optionally be saved as m/z- and Rt-shift-corrected mzXML files.