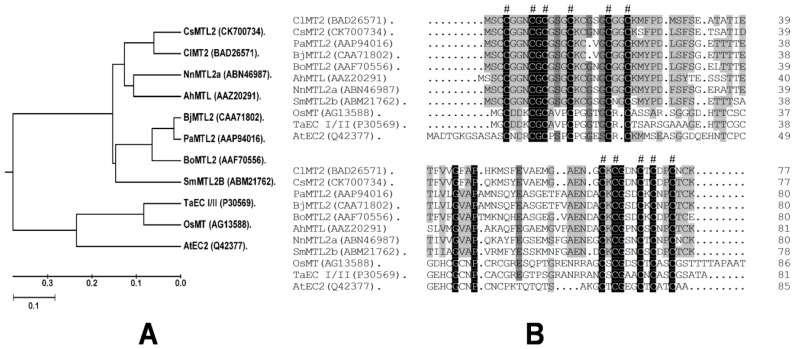
Figure 1.
Phylogenetic tree of 11 selected plant metallothionein (MT) proteins and alignment of the deduced amino acid sequence of Cucumis sativus metallothionein-like 2 (CsMTL2) with MT proteins of other plants. (A) Phylogenic comparison of the deduced amino acid sequences of plant MT proteins. Scale bar denotes 0.1 amino acid substitutions per site. Accession numbers for the MT proteins used are included with the protein name. The MT genes are from Arabidopsis thaliana (At), Triticum aestivum (Ta), Oryza sativa (Os), Brassica juncea (Bj), Brassica oleracea (Bo), Nelumbo nucifera (Nn), watermelon (Cl), Salix matsudana (Sm), Arachis hypogaea (Ah), Pringlea antiscorbutica (Pa), and Cucumis sativus (Cs). (B) Comparison of the deduced amino acid sequence of CsMTL2 with the deduced amino acid sequences of plant MT proteins that have high levels of sequence similarity with CsMTL2. Amino acid residues that are conserved in at least eight of the eleven sequences are shaded, whereas those that are identical in all eleven proteins are in black. The hash symbol represents conserved Cys motifs.