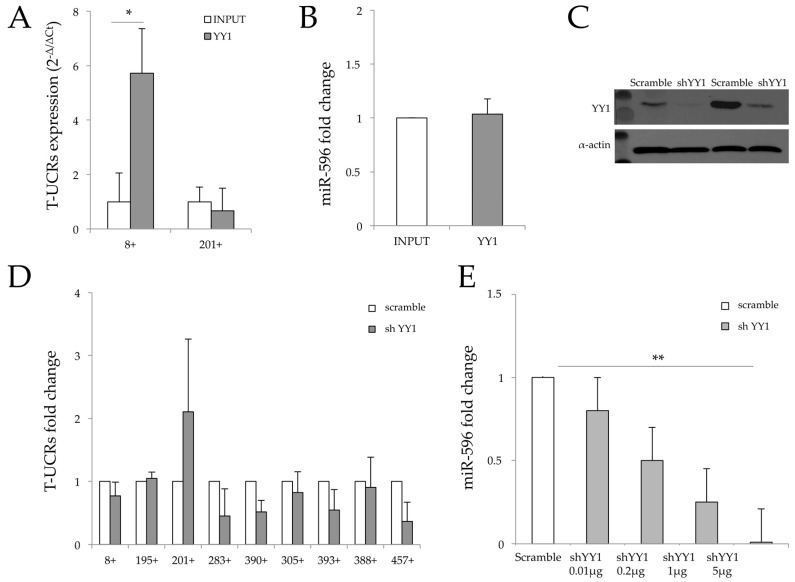
Figure 3.
Role of polycomb YY1 in T-UCR network. (A) RNA-chromatin immunoprecipitation (RNA-ChIP) using YY1 and antibodies followed by real-time PCR (RT-PCR) with T-UCR 8+ and T-UCR 201+ set-primers on pull down materials. Data were expressed as ΔCt of the housekeeping gene U6 and fold change to non-immunoprecipitated samples INPUT (non-immunoprecipitated material) and reported as 2−Δ/ΔCt; (B) RNA-ChIP immunoprecipitation using YY1 antibodies followed by RT-PCR with miR-596 set-primers on pull down materials. Data were expressed as ΔCt of the housekeeping gene U6 and fold change to non-immunoprecipitated samples INPUT (non-immunoprecipitated material); (C) Total protein extracts from J82 cell lines transfected with pbloKit-shYY1 and pbloKit-scramble vectors analysed by Western blot using YY1 antibody. Actin was used as control to equally load the samples; (D) real time PCR (RT-PCR) of the selected T-UCRs, in shYY1-silenced J82 cells compared to empty vector-transfected cells. White boxes indicate cells transfected with pblockiT scramble vector, and grey boxes indicate cells transfected with pblockiT shYY1; (E) qRT-PCR of miR-596 in pblockiT-scramble-transfected cells (white box) and cells transfected with pblockiT shYY1 at different doses as indicated (grey boxes). Data are reported as fold change considering pblockiT-scramble equal to 1 and as mean ± SD of triplicate values. p values were obtained using the Student t-test for three independent samples. * p < 0.05, ** p < 0.01 vs. control.