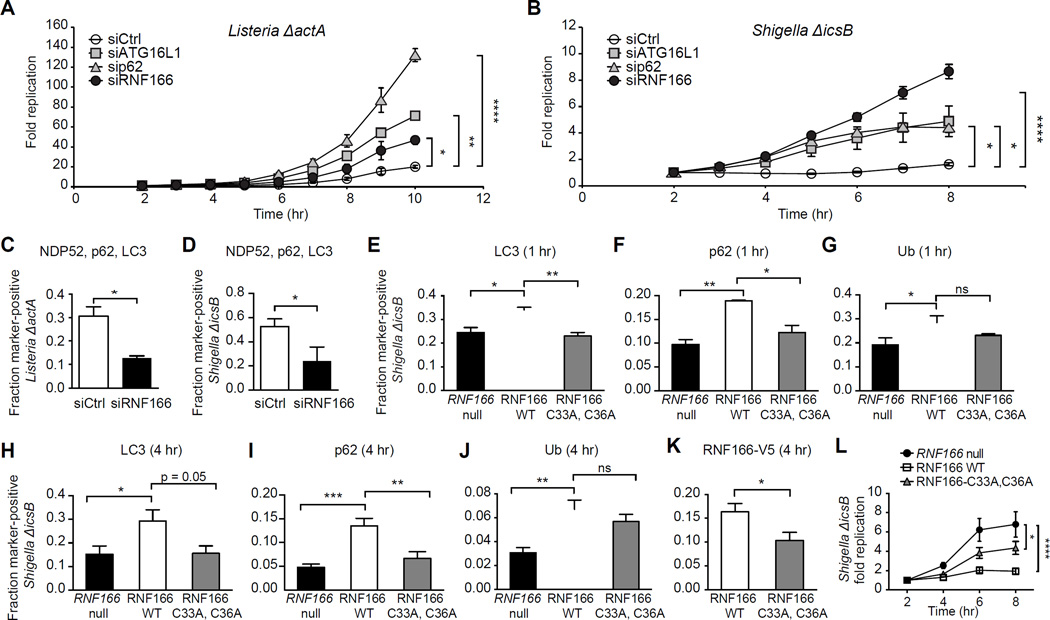
Figure 4. RNF166 Is Required to Inhibit the Intracellular Replication of Listeria and Shigella.
(A) HeLa cells treated with a non-targeting siRNA or siRNA targeting RNF166, p62, or ATG16L1 for 48 hr were infected with Listeria ΔactA expressing luciferase. Cells were treated with gentamicin to remove extracellular bacteria and relative light units were monitored over the indicated time course. Fold replication represents light units over time compared to 2 hr postinfection. Data represent means ± SEM, n = 8.
(B) Cells were treated as in (A) and infected with Shigella ΔicsB expressing luciferase. Data represent means ± SEM, n = 8.
(C, D) HeLa cells were treated with a non-targeting siRNA or siRNA targeting RNF166 for 48 hr, then infected with Listeria ΔactA (C) or Shigella ΔicsB (D) for 1 hr and co-stained for endogenous NDP52, p62, and LC3. The fraction of colocalization of each intracellular bacterium simultaneously with NDP52, p62, and LC3 was scored. > 50 bacteria from three independent experiments were analyzed. Data represent means + SEM.
(E-G) Quantification of LC3 (E), p62 (F), and ubiquitin (G) recruitment to Shigella ΔicsB at 1 hr postinfection in RNF166-null HeLa cells expressing the indicated constructs. Data represent means ± SEM, n = 125 infected cells per group, data pooled from three independent experiments.
(H-K) Quantification of LC3 (H), p62 (I), ubiquitin (J), and RNF166-V5 (K) recruitment to Shigella ΔicsB in RNF166-null HeLa expressing the indicated constructs. Data represent means ± SEM, n = 125 infected cells per group, data pooled from three independent experiments.
(L) Intracellular replication of Shigella ΔicsB expressing luciferase in RNF166-null HeLa cells expressing the indicated constructs. Cells were treated with gentamicin to remove extracellular bacteria and relative light units were monitored over the indicated time course. Fold replication represents light units over time compared to 2 hr postinfection. Data represent means ± SEM, n = 8.
For all panels, *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant. Student’s t test for (C, D, K), one-way ANOVA with multiple comparisons for (A, B, E-J, and L).
See also Figures S3 and S4.