Fig. 2.
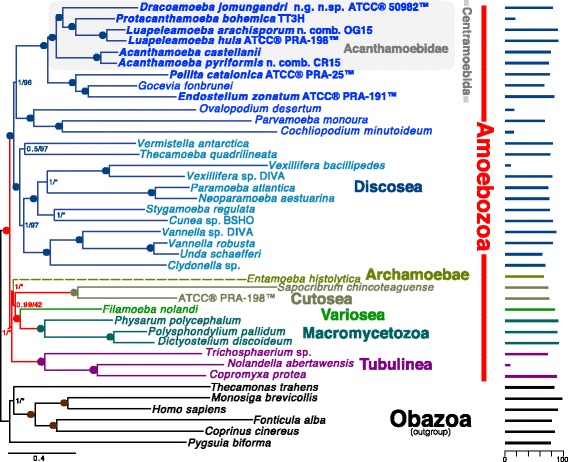
325 gene (102,140 AA sites) phylogeny of Amoebozoa rooted with Obazoa. The tree was built using PhyloBayes-MPI v1.5a under the CAT + GTR model of protein evolution. Values at nodes are posterior probability and ML bootstrap (BS) (1000 ultrafast BS reps, IQ-Tree LG + Γ4 + FMIX(emprical,C20)) values respectively. Circles at nodes represents full support in both analyses (1.0/100). Nodes not recovered in the corresponding ML analysis are represented by an asterisk. The length of the Entamoeba branch is shown as a dashed line to represent that its total length has been reduced by 50%. Bars along the right side of the figure show the percent of the total data set available for each taxon. Novel data was generated in this study for taxa whose names are bold
