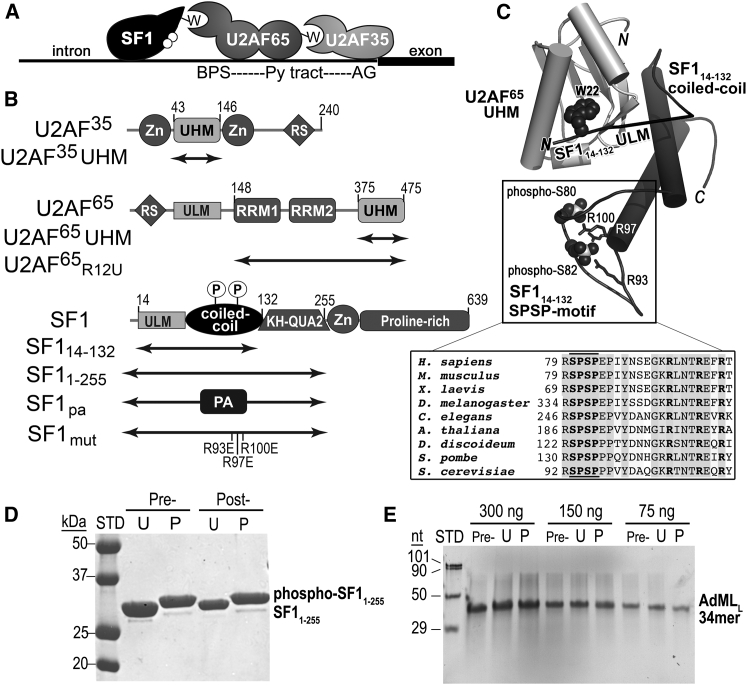
Figure 1.
SF1 structure and function in pre-mRNA splicing. (A) Schematic diagram of the early-stage SF1-U2AF65-U2AF35 splicing-factor complex recognizing the 3′ splice site. ULMs are represented by “W.” White circles represent UHMK1-phosphorylation sites. BPS, branch point sequence; Py, polypyrimidine. (B) Domains of SF1, U2AF65, and U2AF35 splicing factors. The boundaries of constructs used in this study are indicated by double-headed arrows. Zn, CCCH zinc knuckle; UHM, U2AF homology motif; ULM, U2AF ligand motif; RRM, RNA recognition motif, RS, arginine-serine repeat; KH-QUA2, K homology with quaking-2 motif; PA, protein A; circled P, UHMK1-phosphorylation sites. (C) Structure of the phosphorylated SF114–132-U2AF65 UHM complex (PDB: 4FXW). A line representing residues that are absent from the electron density connects the C-terminus of the ULM to the coiled-coil motifs. The tryptophan of the ULM (W22) and phosphorylated serines are shown as spheres and arginine residues that are mutated in this study are shown in stick representation. The inset shows a sequence alignment of the UHMK1-phosphorylated SPSP motif (bold (between lines)). (D and E) Samples of (D) phospho-SF11–255 and (E) AdMLL RNA under experimental conditions. Samples either before (Pre-) or after (Post-) incubating at 23°C for 2 h in the presence of a 10-fold excess of SF1 (U) or phospho-SF1 (P) were analyzed by denaturing gel electrophoresis and stained using (D) Coomassie blue or (E) SYBR Gold, respectively. The phosphorylated form of SF11–255 runs higher by SDS-PAGE. STD, size markers; nt; nucleotides. Conditions were identical to those of fluorescence assays with the exception that the RNase inhibitor Superase-In (Ambion, Life Technologies) was omitted here to fully rule out RNase contamination.