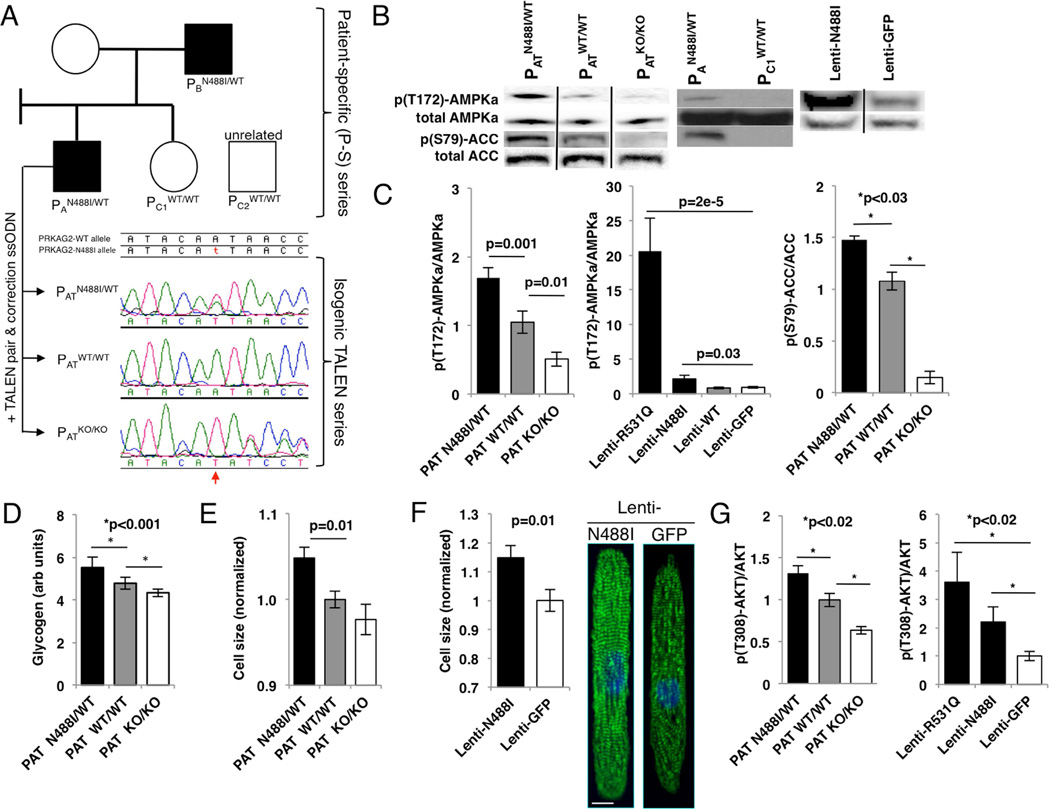
Figure 1. PRKAG2 cardiomyopathy iPS-CMs recapitulate hypertrophy and glycogen accumulation due to AMPK activation.
(A) IPScs were engineered from two affected individuals (PAN488I/WT and PBN488I/WT), and a related (PC1WT/WT) and unrelated control (PC2WT/WT) (circle=female, square=male; shaded=PRKAG2 cardiomyopathy, unshaded=normal heart). PAN488I/WT iPScs were genome-edited with TALENs and a wildtype PRKAG2 oligonucleotide to create an isogenic series at the N488I locus (PATN488I/WT, PATWT/WT and PATKO/KO). Sanger tracings of PRKAG2 amplicons derived from the isogenic TALEN series (red arrow=A/T substitution). (B) Representative immunoblots probed with anti-p(T172)-AMPKα subunit, p(S79)-ACC and total AMPKα and ACC, and (C) quantified by densitometric analysis (n≥3). (D) Quantification of intracellular glycogen in iPS-CMs (n≥3). (E) iPS-CM size measured by normalized forward scatter (FSC) by flow cytometry (n≥15 differentiations) and by (F) pixel area on fibronectin lines (n≥20 myocytes); representative myocytes stained with anti-cardiac actin-in A (green) and DAPI (blue; scale bar=10 microns). (G) Quantification of anti-p(T308)-AKT by normalized densitometry (n≥3 lanes each) of immunoblots from lysates derived from iPS-CMs. Significance assessed by Student’s t-test (C–G) and error bars are mean +/− SEM (C–G).