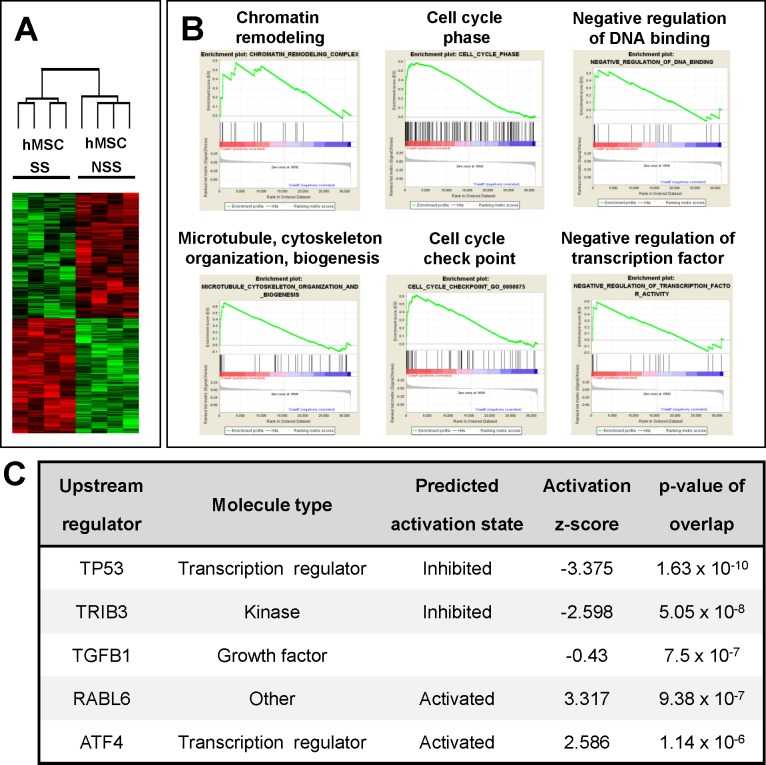
Fig 7. Distinct signaling pathways of the MSCs under stimulatory and non-stimulatory conditions.
Three independent lines of hMSCs cultured under different conditions were subjected to microarray analysis for transcriptomic differences. Transcriptomes exhibiting significant difference (p<0.01) between the MSCs were subjected to gene set enrichment analysis (GSEA) and pathway analysis (Ingenuity pathway analysis; IPA). Shown are the plots of microarray (A) and representative enrichment profiles from GSEA (B). Full list of GOs are shown in S1 Table. (C) Distinct upstream signals of MSCs identified by pathway analysis. Five candidate upstream signals exhibiting significant alterations in the MSCs under stimulatory conditions relative to the MSCs under non-stimulatory conditions are shown.