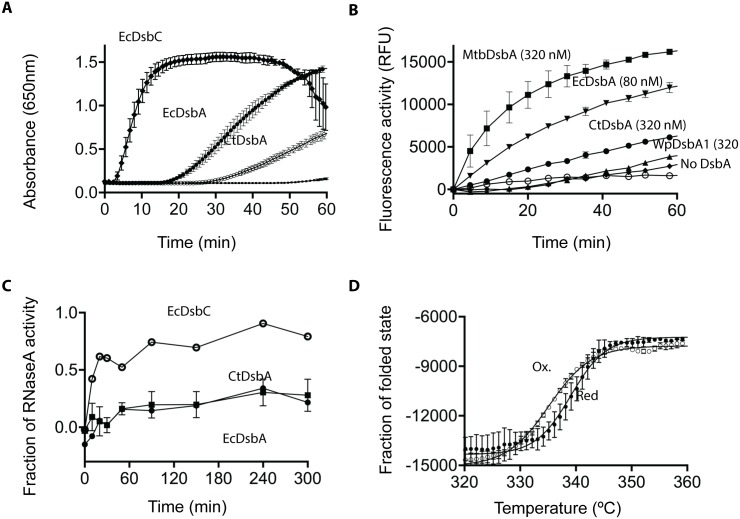
Fig 1. Biochemical characterization of CtDsbA.
A Reduction of insulin (131 μM) was measured as increase in absorbance at 650nm in 0.1mM sodium phosphate buffer, pH 7, 2mM EDTA. The reaction was performed in the absence (■) or presence of 10 μM EcDsbC (●), EcDsbA (♦) or CtDsbA (○). Representative data are shown for the absence and presence of 10 μM EcDsbA. Mean and SD are shown for two biological replicates (three biological replicates for CtDsbA). B 80 nM EcDsbA (▼) and 320 nM CtDsbA (●), MtbDsbA (■) and WpDsbA1 (▲) oxidize a fluorescently labeled protein in the presence of 2 mM GSSG. GSSG shows only limited oxidizing activity in the absence of a DsbA protein (■). The buffer only control (○) shows no oxidizing activity. For MtbDsbA, WpDsbA1, EcDsbA and CtDSbA mean and SD of two biological replicates are shown (for each biological replicates four technical replicates was performed.) For the buffer and GSSG only controls, mean and sd for four technical replicates are shown. C Isomerase activity was assessed as the ability to refold scrambled RNAseA. ScRNase (40 μM) was incubated in 0.1 M sodium phosphate buffer pH 7.0, 1 mM EDTA, 10 μM DTT in the absence and presence of 10 μM EcDsbA (■), EcDsbC (○) and CtDsbA (●). RNase activity was monitored as the cleavage of cCMP which leads to an increase in absorbance at 296 nm. Mean and SD for three biological replicates is shown for CtDsbA. EcDsbC and EcDsbA is able to restore ~80% and ~20% of RNaseA activity, which is equivalent to what has been reported previously [8] D Temperature induced unfolding of oxidized (○) and reduced (●) CtDsbA was determined by far UV CD spectroscopy. The thermal unfolding of CtDsbA results in an increase in molar ellipticity at 220 nm showing that the reduced form of CtDsbA is more stable than the oxidized form. Mean and SD are shown for two biological replicates.