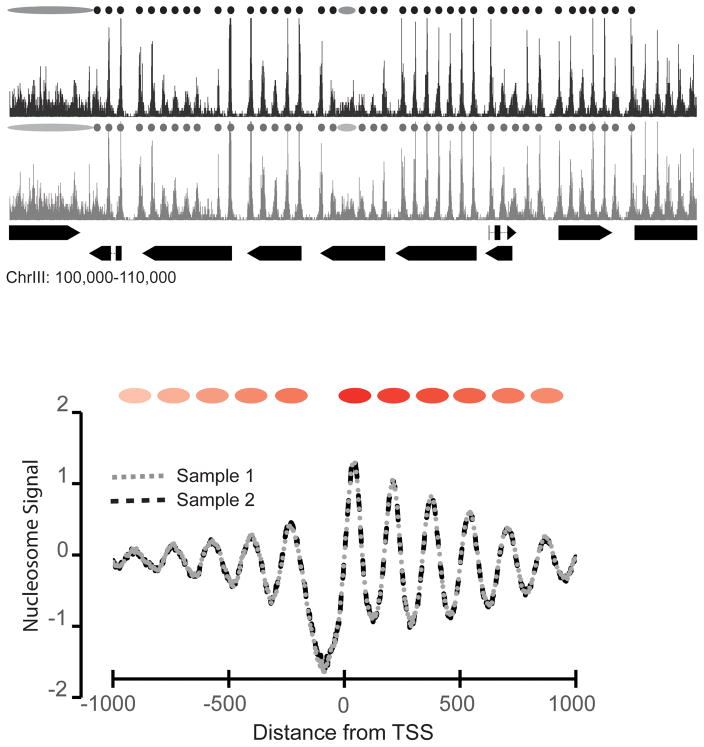
Figure 1.
Nucleosome positions are highly reproducible in S. cerevisiae cells. Top: Two representative genome browser images showing nucleosome dyads from MNase-seq experiments in independent S. cerevisiae isolates. Small circles denote well-positioned nucleosomes while large ovals represent poorly-positioned nucleosomes. Pointed rectangles denote annotated transcription units. Bottom: Overlay of nucleosome dyad signal at transcription start sites (TSS) for the two isolates in (from top). Red ovals represent positioned nucleosomes with respect to the TSS. Data was obtained from Gene Expression Omnibus (GEO) GSE72572 [27].