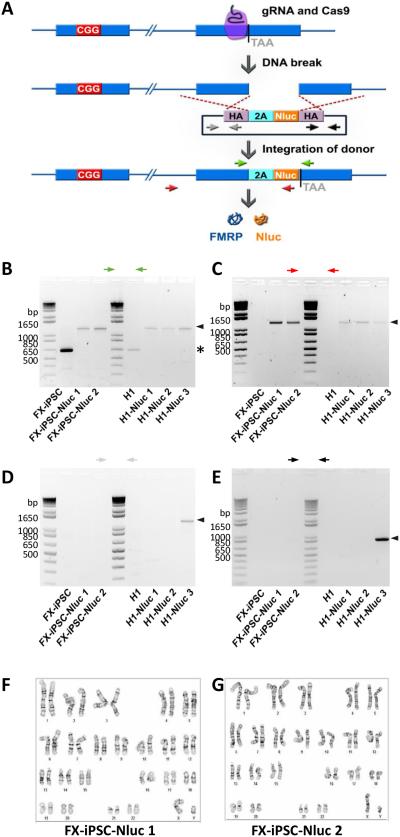
Figure 1. Generation of FMR1-Nluc reporter cell lines from H1 ESCs and FXS-iPSCs.
(A) Generation of reporter lines by using CRISPR/Cas9-mediated genome editing. Cleavage of FMR1 DNA by a guide RNA (gRNA)-Cas9 complex was followed by homologous recombination with the homology arms (HA) in a donor plasmid containing Nano luciferase gene (Nluc) and the 3’ coding sequence of FMR1 gene. (B, C) PCR validation of expected donor integration through homologous recombination in reporter lines. One pair of primers (green arrows) amplified the region of FMR1 gene with Nluc insertion yielding difference sizes from parental or targeted alleles. Another pair of primers (red) detected only targeted allele, but not parental allele. Black Arrowheads indicate expected bands of reporter lines and asterisk indicates bands of parental lines. (D, E) PCR validation of the absence of random integration of donor plasmid in reporter lines. FX-iPSC-Nluc 1 and 2 and H1-Nluc 1 and 2 were negative while H1-Nluc 3 was positive for random inserted donor plasmid. Black Arrowheads indicate expected bands from random, non-homology directed integration of the donor plasmid. (F, G) Karyotypes of FX-iPSC-Nluc 1 (F) and FX-iPSC-Nluc 2 (G). Normal 46XY karyotypes were observed.