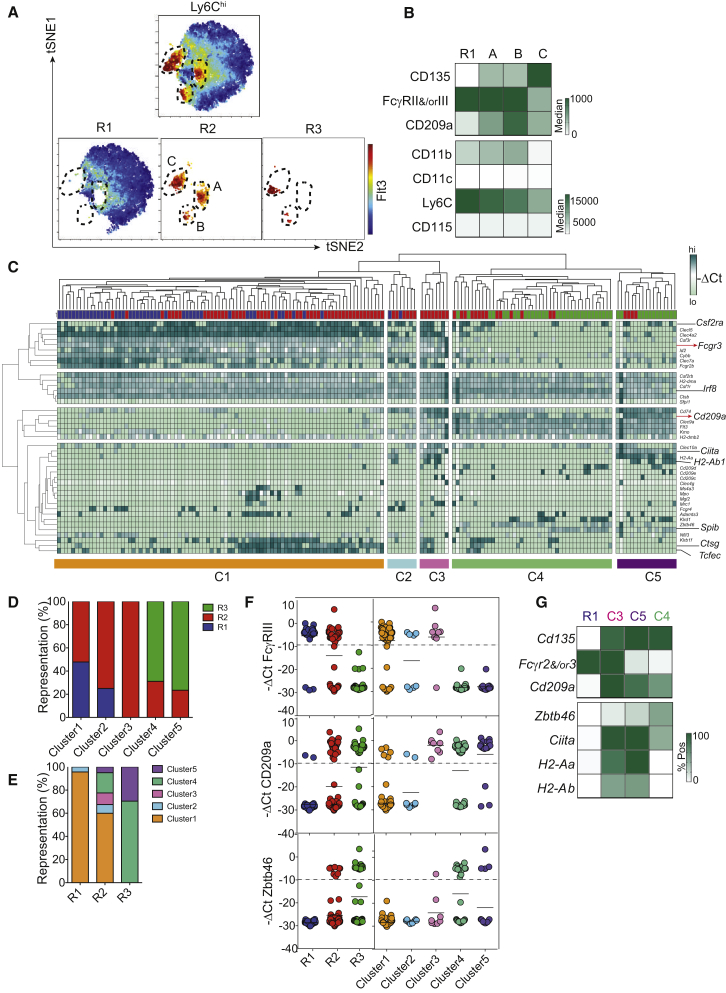
Figure 2.
Transcriptional Profiling of Monocyte Subsets R1 and R2
(A) Nonlinear dimensionality reduction analysis of Ly6ChiCD115+cells. t-SNE maps of total Lin−MHCII−CD115+SIRPα+ cells, R1 and R2 monocytes, and R3 pre-DCs are based on the parameters CD115, SIRPα, Ly6C, Flt3, CD11c, CD209a, and FcγRII and/or FcγRIII. Color scale indicates Flt3 expression.
(B) Expression analysis of t-SNE-generated sub-populations of R2 and expression of Flt3, FcγRII and/or FcγRIII, CD209a, CD11b, CD11c, Ly6C, and CD115 of t-SNE-generated Flt3+ populations A–C with R1 monocytes. CD209a expression is shown as the difference between MFI and fluorescence minus one (FMO) control for all four populations.
(C) Heterogeneity in population R2. The hierarchical clustering dendogram (top and left margins) is based on −ΔCt values from single-cell multiplex qPCR analysis of 44 R1 (blue), 81 R2 (red), and 44 R3 (green) single cells for 42 genes.
(D) Representation of populations R1–R3 within the five clusters defined in (C).
(E) Representation of the five clusters within populations R1–R3.
(F) Single-cell expression of Fcgr3, CD209a, and Zbtb46 in populations R1–R3 or clusters C1–C5. Each dot represents the −ΔCt value of a single cell.
(G) Analysis of the defining genes of population R1 with clusters C3–C5. The heatmap compares mRNA expression of MHCII-related genes Cd135, Zbtb46, Cd209a, and Fcgr2 and/or Fcgr3 on single cells between clusters C3–C5 and monocyte population R1.
Please also refer to Figure S2.