ABSTRACT
The multisubunit cation/proton antiporter 3 family, also called Mrp, is widely distributed in all three phylogenetic domains (Eukarya, Bacteria, and Archaea). Investigations have focused on Mrp complexes from the domain Bacteria to the exclusion of Archaea, with a consensus emerging that all seven subunits are required for Na+/H+ antiport activity. The MrpA subunit from the MrpABCDEFG Na+/H+ antiporter complex of the archaeon Methanosarcina acetivorans was produced in antiporter-deficient Escherichia coli strains EP432 and KNabc and biochemically characterized to determine the role of MrpA in the complex. Both strains containing MrpA grew in the presence of up to 500 mM NaCl and pH values up to 11.0 with no added NaCl. Everted vesicles from the strains containing MrpA were able to generate a NADH-dependent pH gradient (ΔpH), which was abated by the addition of monovalent cations. The apparent Km values for Na+ and Li+ were similar and ranged from 31 to 63 mM, whereas activity was too low to determine the apparent Km for K+. Optimum activity was obtained between pH 7.0 and 8.0. Homology molecular modeling identified two half-closed symmetry-related ion translocation channels that are linked, forming a continuous path from the cytoplasm to the periplasm, analogous to the NuoL subunit of complex I. Bioinformatics analyses revealed genes encoding homologs of MrpABCDEFG in metabolically diverse methane-producing species. Overall, the results advance the biochemical, evolutionary, and physiological understanding of Mrp complexes that extends to the domain Archaea.
IMPORTANCE The work is the first reported characterization of an Mrp complex from the domain Archaea, specifically methanogens, for which Mrp is important for acetotrophic growth. The results show that the MrpA subunit is essential for antiport activity and, importantly, that not all seven subunits are required, which challenges current dogma for Mrp complexes from the domain Bacteria. A mechanism is proposed in which an MrpAD subcomplex catalyzes Na+/H+ antiport independent of an MrpBCEFG subcomplex, although the activity of the former is modulated by the latter. Properties of MrpA strengthen proposals that the Mrp complex is of ancient origin and that subunits were recruited to evolve the ancestral complex I. Finally, bioinformatics analyses indicate that Mrp complexes function in diverse methanogenic pathways.
KEYWORDS: Methanosarcina, methanogen, sodium/proton antiport
INTRODUCTION
Cation/proton antiporters are of fundamental importance and are widely distributed in organisms from all three phylogenetic domains (Bacteria, Archaea, and Eukarya) of life. Antiporters in the domains Bacteria and Archaea are secondary transporters driven by an electrochemical gradient of protons (high outside) to achieve cytoplasmic homeostasis of cations. Most cation/proton antiporters are monomers, in contrast to the cation/proton antiporter 3 (CPA3) family, also called Mrp complexes, which are comprised of six or seven subunits, depending on whether subunits MrpA and MrpB are fused (1–3). Analyses of genomic sequences show Mrp complexes are broadly distributed among species with diverse physiologies from the domains Bacteria and Archaea, although investigations have focused on Bacteria to the exclusion of Archaea. Mrp complexes from Bacteria have been implicated in functions that include Na+/H+ antiport (2–8), K+/H+ antiport (9), pH homeostasis (7, 10, 11), resistance to bile salts (7, 12), arsenite oxidation (8), pathogenesis (12, 13), and energy conversion (14, 15). Mrp complexes and their subunits are anticipated to have undiscovered functions (1). A complex containing both Mrp and hydrogenase (Mbh) subunits is present in anaerobic microbes that oxidize ferredoxin and produce H2 coupled with generation of a proton gradient that drives ATP synthesis for growth (16). Analysis of available genome sequences revealed homologs of Mrp-Mbh complexes in the Bacteria and Archaea. The biological diversity and evolutionary importance of Mrp complexes in nature is underscored by the resemblance of MrpA, MrpD, and MrpC to subunits of the coenzyme F420 dehydrogenase complex from methane-producing species in the domain Archaea (17).
MrpA and MrpD are homologous to the antiporter-like subunits NuoL and NuoM/NuoN, respectively, of respiratory complex I from the domains Bacteria and Eukarya (18–20). Indeed, NuoL and NuoN are functional replacements for MrpA and MrpD of Bacillus species (20). The recently reported crystal structure of complex I highlights the similarity of MrpA and MrpD from the domain Bacteria with antiporter-like subunits NuoL and NuoM/NuoN, respectively, of complex I (21). Indeed, it has been proposed that subunits of the Mrp complex were recruited to evolve the ancestral complex I (22). This proposal is also consistent with the proposal that the Mrp complex was a component of the ancient anaerobic core present in the ancestor of the bacterial and archaeal lineages (23).
Understanding the antiport mechanism of Mrp complexes has been hindered by the lack of a crystal structure and the subunit multiplicity. The seven-subunit MrpABCDEFG complexes from Bacillus species have been model systems for investigation, with a focus on the MrpA and MrpD subunits that contain transmembrane acidic residues, consistent with ion translocation functions (24). Although MrpA and MrpD subunits from the domain Bacteria are implicated in translocation of Na+ and H+, respectively, a consensus has emerged that all seven subunits are required for antiport activity (2, 3, 11, 20, 25, 26). Specifically, expression of an MrpAD subcomplex in the antiporter-deficient Escherichia coli strain KNabc showed no antiport activity (27). Furthermore, MrpA and MrpD were reported to be inactive when expressed alone (1). On the other hand, a fused MrpA-MrpD homolog in the alkaliphilic Natranaerobius thermophilus displayed Na+/H+ antiport activity when produced in E. coli strain KNabc (5, 28). Clearly, further research is necessary to clarify the mechanism and multifaceted roles of Mrp complexes in diverse species.
Even though the sequenced genomes of species in the domain Archaea are annotated with genes encoding Mrp complexes, the only reported investigation is that for the seven-subunit MrpABCDEFG complex from the methane-producing (methanogen) Methanosarcina acetivorans (15). Comparisons of the wild type versus a ΔmrpA mutant show that the Mrp complex is essential for optimal ATP synthesis and growth of cells converting acetate to methane and carbon dioxide (15). Growth with acetate generates both Na+ and H+ gradients that drive ATP synthesis by an ATP synthase dependent on both Na+ and H+ gradients (29). It is postulated that the Mrp complex from M. acetivorans functions to adjust the Na+/H+ ratio optimally for ATP synthase (15). Here, we report properties of the MrpA subunit of M. acetivorans produced in E. coli strains with Na+/H+ antiporter deficiencies that establishes a role for MrpA in the MrpABCDEFG complex important for acetotrophic growth.
RESULTS AND DISCUSSION
Transformation with mrpA from M. acetivorans complements the Na+ and alkali sensitivities of antiporter-deficient E. coli strains.
Antiporter-deficient E. coli strains have been used extensively to evaluate the Na+/H+ antiport activity of Mrp complexes and subcomplexes (4–6, 12, 27). The antiporter-deficient strains EP432 and KNabc are highly sensitive to growth inhibition by NaCl concentrations greater than 200 mM compared to the E. coli wild type, as confirmed by the results shown in Fig. 1A. Both the E. coli mrpA EP432 and E. coli mrpA KNabc transformants showed striking Na+ resistance up to 500 mM. Strains EP432 and KNabc are also sensitive to pH values above 8.0 (Fig. 1B). Both transformants supported higher final growth yields than the antiporter-deficient strains at pH values up to 11.0 in the absence of added Na+ (Fig. 1B). To our knowledge, this is the first report showing that MrpA from any source participates in Na+/H+ antiport activity that is independent of an intact MrpABCDEFG complex.
FIG 1.
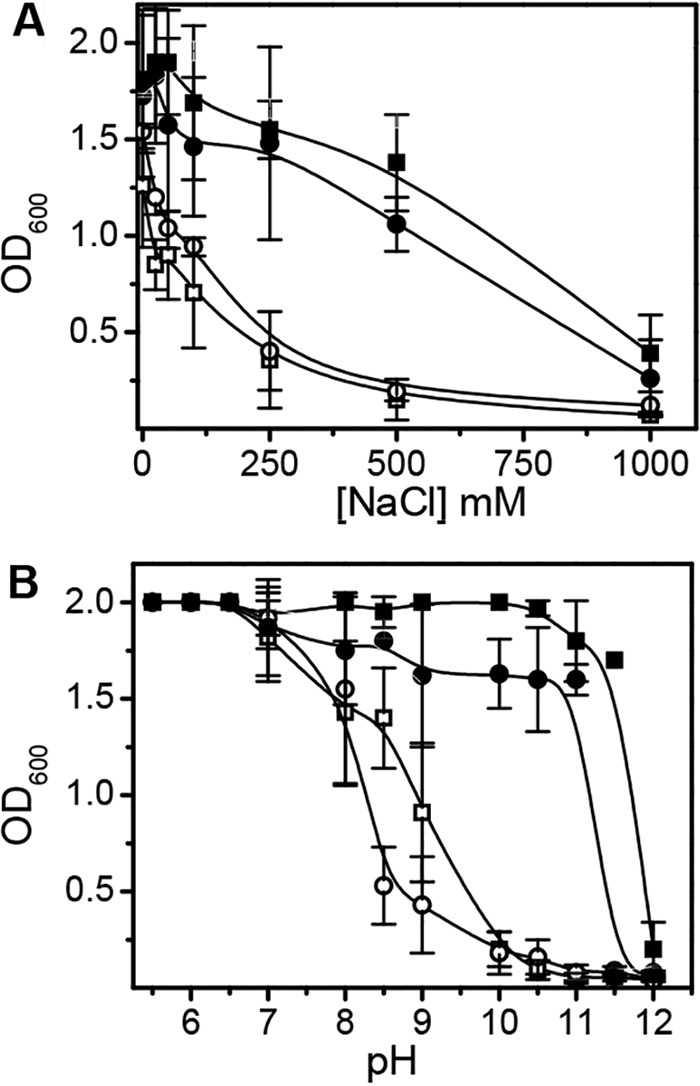
Effects of NaCl and pH on growth of transformed E. coli strains mrpA EP432 and mrpA KNabc. (A) Transformants were grown at pH 7 in media containing the indicated concentrations of NaCl. (B) Transformants were grown with no added NaCl at the indicated pH values. The media contained approximately 12 mM residual NaCl. □, strain EP432; ○, strain KNabc; ■, transformed strain mrpA EP432; ●, transformed strain mrpA KNabc. The cultures were grown for 10 to 11 h. The values are the means of the results of at least 4 experiments ± SD.
MrpA catalyzes secondary Na+ (Li+)/H+ antiport with a high apparent Km for Na+ and a neutral pH optimum.
Antiport was assessed by energization of everted (inside out) membrane vesicles with the addition of an electron donor that elicits a pH gradient (ΔpH; acid inside) in the vesicles. The ΔpH was monitored by quenching of acridine orange fluorescence until the steady state is reached. Activity was evaluated by the percent dequenching that results from addition of the monovalent cation required for antiport.
Everted vesicles were able to form a ΔpH after the addition of d-lactate (data not shown), although more reliable results were attained with NADH, where up to 90% of total dequenching determined by carbonyl cyanide m-chlorophenylhydrazone (CCCP) addition was achieved (Fig. 2A to C), indicating high-quality preparations of sealed everted vesicles; hence, NADH was used in all the experiments. Figure 2A to C shows representative traces of ΔpH formation in everted vesicles from E. coli strain K-12 and the transformed strains mrpA EP432 and mrpA KNabc at pH 8.0. The H+ gradient was abated almost completely by addition of NaCl, as judged by CCCP addition, a result indicating that the H+ gradient was exhausted by antiport with Na+ dependent on MrpA. For comparison, the H+ gradient formed by membranes from Na+-sensitive strain EP432 was not abated by NaCl addition, attributable to the absence of native Na+/H+ antiporters in the strain (Fig. 2D). These data indicate that the MrpA subunit from M. acetivorans was synthesized and correctly oriented in the membranes of E. coli strains, necessary for Na+/H+ antiport activity. The results reinforce those shown in Fig. 1, supporting the conclusion that MrpA is essential for Na+/H+ antiport activity. The results further show that activity is independent of an intact MrpABCDEFG complex, contrary to previous assertions that intact complexes from the domain Bacteria are required for activity (2, 3, 11, 20, 25, 26).
FIG 2.
Antiport activity of M. acetivorans MrpA in everted membrane vesicles of E. coli strains. The assay mixtures contained 50 to 75 μg of vesicle protein in 2 ml of 50 mM Bis-Tris propane buffer at pH 8.0. (A) Strain K-12. (B) Strain mrpA EP432. (C) Strain mrpA KNabc. (D) Strain EP432. The traces are representative of the results of 4 independent experiments. The arrows indicate (i) the time scale (50s), (ii) when respiration was initiated with 0.5 mM NADH (NADH), (iii) when quenching reached steady state and a nonsaturating 50 mM NaCl was added (NaCl), and (iv) when 5 μM CCCP was added (CCCP). ΔF, relative change in fluorescence.
The concentration of ion producing half-maximal dequenching is a good estimate of the apparent Km of sodium/proton antiporters and is used routinely (4). Nonlinear fitting of activities determined with mrpA EP432 vesicles over a range of concentrations up to 150 mM resulted in apparent Km values for Na+ and Li+ of 47 ± 16 and 45 ± 14 mM (mean ± standard deviation [SD]; n = 4), respectively (Fig. 3A and B). Lineweaver-Burk plots of the data (Fig. 3A and B, insets) yielded apparent Km values for Na+ and Li+ of 31 and 63 mM. Dequenching reached 80 and 90% of maximum with Na+ and Li+, respectively (Fig. 3). Activities with K+ were low and unreliable, reaching only 20% of maximum dequenching of fluorescence, which precluded a reliable evaluation of the apparent Km. The low activity with K+ compared with Na+ and Li+ shows that Na+ is the preferred physiological cation, similar to what has been reported for complete Mrp complexes from Staphylococcus aureus, Bacillus sp., and Vibrio cholerae of the domain Bacteria (4, 12). Remarkably, the apparent Km values determined for Na+ and Li+ with E. coli mrpA EP432 vesicles were more than 400-fold higher than the Km values reported for E. coli EP432 vesicles containing Mrp complexes from species in the domain Bacteria that function in resistance to salt and alkaline pH (4, 12). It could be argued that the MrpA subunit has considerably lower affinities for Na+ when it is present in the complete Mrp complex, consistent with the proposal that Mrp complexes are a consortium of antiporters that function in concert (1). However, the differences in apparent cation affinities may also reflect a role for the M. acetivorans Mrp complex in adjusting the ratio of the Na+ and H+ gradients optimally for Na+- and H+-dependent ATP synthase (15, 29).
FIG 3.
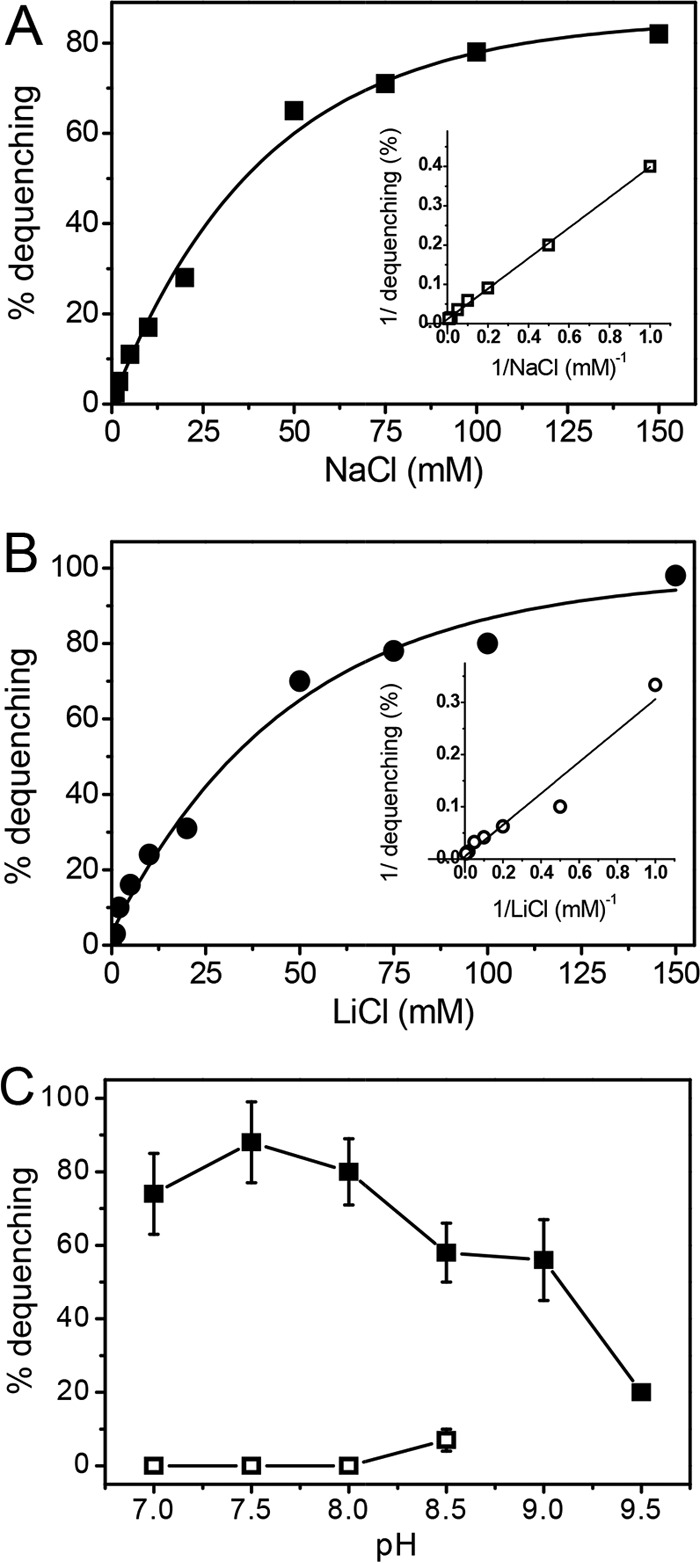
Antiport activity as a function of the cation concentration and pH. (A and B) Fluorescence-based assays of the Na+/H+ antiport activities of MrpA in E. coli strain mrpA EP432 vesicles were conducted at pH 8.0 over a range of concentrations of added NaCl (A) and LiCl (B). (C) Effect of pH on Na+/H+ antiporter activity of MrpA in vesicles of E. coli strain EP432 (□) and E. coli strain mrpA EP432 (■). The assay mixtures contained 100 mM NaCl. The error bars indicate standard deviations of the results of three independent assays.
The Na+/H+ antiport activity profile as a function of pH was determined with E. coli mrpA EP432 vesicles energized with NADH (Fig. 3C). Similar results were obtained when d-lactate replaced NADH (not shown). Optimum activity was obtained between pH 7.0 and 8.0, with significant activity maintained up to pH 9.0. Activity was barely detectable with control E. coli EP432 vesicles. The results are in contrast to the optimal pH activity between 8.0 and 9.5 reported for E. coli EP432 vesicles containing complete Mrp complexes from Bacillus sp. and V. cholerae (4, 12). Conversely, the results shown in Fig. 3C are similar to those reported for E. coli EP432 vesicles containing the complete Mrp complex from S. aureus (4). Nonetheless, the difference in pH profiles of antiporter activity between M. acetivorans and species from the domain Bacteria, combined with the different affinities for Na+, suggests a different function for the Mrp complex from M. acetivorans, consistent with the previous proposal that the complex functions primarily in energy conversion (15).
Bioinformatics analyses.
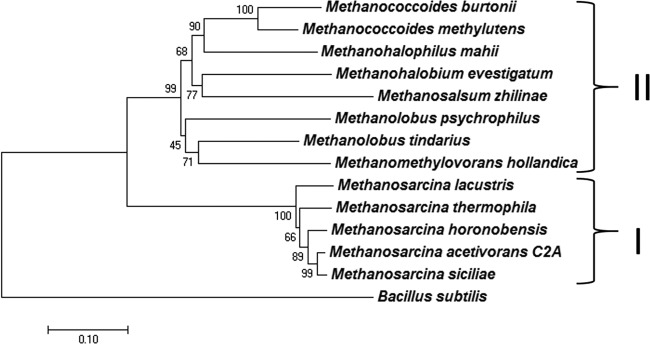
A BLASTp search of databases (http://www.ncbi.nlm.nih.gov/pubmed) was conducted with M. acetivorans MrpA (gene locus identifier MA4572) as the query. Of the 100 significant alignments retrieved, homologs with the greatest identity (56 to 98%) were from methanogen species, followed by homologs (38 to 43% identity) from Bacillus species. The methanogen homologs were divisible into two clades (Fig. 4). Clade I contained sequences (91 to 98% identity) from acetate-utilizing Methanosarcina species, except for the freshwater counterparts Methanosarcina mazei and Methanosarcina barkeri, whereas clade II was comprised of sequences (56 to 62% identity) from diverse genera that utilize only methylotrophic substrates (methanol, methylamines, and dimethyl sulfide) for growth and methanogenesis. An alignment of sequences from clades I and II showed two consensus sequences present in clade I (SETY and GIGVG), with unknown function, that are absent in all the sequences of clade II. The MrpA homologs from Methanococcoides burtonii and Methanococcoides methylutens in clade II were adjacent to a contiguous set of genes encoding homologs of the remaining six subunits of the canonical seven-subunit Mrp complex. Although gene context data were not available for other species in clade II, the results are consistent with a role for Mrp complexes in species from both clades. A role for the Mrp complex in M. acetivorans was previously proposed, where the complex adjusts the ratio of Na+ and H+ gradients optimally for driving ATP synthesis by sodium- and proton-dependent ATP synthase during growth with acetate (15). The genomes of M. burtonii and M. methylutens in clade II are annotated with genes encoding subunits of the canonical membrane-bound Rnf complex shown to pump Na+ during electron transport in M. acetivorans (30, 31). The Rnf complex in these methylotrophic methanogens has the potential for participation in the membrane-bound electron transport chain that also generates a H+ gradient, in addition to a Na+ gradient (32). Thus, the Mrp complex may also function to adjust the ratio of Na+ and H+ gradients optimally for driving ATP synthesis, although it would depend on a sodium- and proton-dependent ATP synthase in methylotrophic methanogens that has not been reported. Interestingly, it has been speculated that differences in the abundances of Mrp subunits during growth of M. burtonii with methanol at different temperatures reflect a role for the Mrp complex in energy conservation (33). On the other hand, the differences that separate clades I and II possibly indicate different functions. Indeed, except for the freshwater strains Methanomethylovorans hollandica and Methanolobus psychrophilus, species in clade II were isolated from marine environments, consistent with a potential role for the Mrp complexes in maintaining salt homeostasis.
FIG 4.
Distance tree of MrpA from M. acetivorans, MrpA homologs from other methanogens, and MrpA from Bacillus subtilis. The evolutionary history was inferred using the neighbor-joining method with significant alignments (see Fig. S5 in the supplemental material) recovered from the NCBI database queried with MrpA (MA4572; MA_RS23835) from M. acetivorans (53). The optimal tree, with a sum of branch lengths equal to 2.3, is shown. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches (54). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in units of the number of amino acid substitutions per site (55). All positions containing gaps and missing data were eliminated. There were a total of 776 positions in the final data set. Evolutionary analyses were conducted in MEGA7 (56). The percent identities relative to MrpA from M. acetivorans (shown in parentheses) were as follows: WP_011024445.1, Methanosarcina acetivorans (100%); WP_048174247.1, Methanosarcina siciliae (98%); WP_048136646.1, Methanosarcina horonobensis (96%); WP_048125080.1, Methanosarcina thermophila (93%); WP_048124368.1, Methanosarcina lacustris (91%); WP_023846288.1, Methanolobus tindarius (63%); WP_013194388.1, Methanohalobium evestigatum (59%); WP_048204897.1, Methanococcoides methylutens (62%); WP_013037209.1, Methanohalophilus mahii (60%); WP_015325011.1, Methanomethylovorans hollandica (61%); WP_048147233.1, Methanolobus psychrophilus (59%); WP_011498313.1, Methanococcoides burtonii (59%); WP_013898326.1, Methanosalsum zhilinae (56%); WP_014470898.1, Bacillus subtilis (40%). I and II, clades I and II.
Archaeal MrpA subunits from M. acetivorans and methylotrophic methanogens, represented by M. burtonii, are formed by two transmembrane-rich domains, similar to MrpA from Bacillu subtilis representing the domain Bacteria (1, 34) (see Fig. S4 in the supplemental material). The N-terminal domains of archaeal and B. subtilis MrpA subunits (see Fig. S4A in the supplemental material) show similarity to NuoL of complex I from E. coli. However, the C-terminal domains of archaeal and B. subtilis MrpA subunits (see Fig. S4B in the supplemental material) do not share significant identity with subunits from complex I of E. coli. Residues essential for functionality in MrpA from B. subtilis and NuoL from E. coli (E141, K227, H252, and K402) (18, 34–38) are conserved in the archaeal MrpA subunit, suggesting a role for the residues in ion transport. Residues directly involved in proton translocation of antiporter-like subunits in complex I are conserved in MrpA of M. acetivorans (E141 and K227) (18). A section in the piston region of NuoL from E. coli complex I containing the important residues D542 and D546 and the essential D563 (39) is deleted in the piston region of archaeal and B. subtilis MrpA subunits.
Overall, the sequence comparisons indicate that MrpA subunits from the domains Archaea and Bacteria share an ancestor, although they have evolved somewhat different functions. The recent proposal that conversion of acetate to methane in M. acetivorans evolved by way of horizontal gene transfer suggests the possibility that MrpA of M. acetivorans was acquired similarly and then evolved for a specific function (40). Indeed, nearly one-third of the open reading frames (ORFs) in the genome of M. mazei have the greatest identity to homologs in the domain Bacteria (41). Likewise, genome analyses also indicate a major role for horizontal gene transfer in the evolution of the methylotrophic methanogen M. burtonii (42).
Homology molecular model of MrpA.
The N-terminal domain of MrpA (MrpA_N) is homologous to the first 14 transmembrane sequences (TMS) of NuoL, and the C-terminal domain (Mrp_C) is homologous to the five TMS of NuoJ of E. coli complex I, the crystal structure of which is known (35). Additionally, MrpA from M. acetivorans is homologous to MrpA from B. subtilis, in which the N-terminal domain replaces the function of NuoL and the C-terminal domain replaces the function of NuoJ (see Fig. S4 in the supplemental material) (25, 34). Thus, MrpA_N and MrpA_C were modeled independently using the corresponding NuoL and NuoJ templates.
Figure 5 shows a tentative model of MrpA from M. acetivorans built with the MrpA_N and MrpA_C models using a template constructed from the NuoLNMJ subunits of E. coli complex I. The model shows an arrangement in which Mrp_N corresponds to NuoL and Mrp_C corresponds to NuoJ (21). NuoL contains a C-terminal extension called the piston that interfaces with NuoJ in the crystal structure of complex I. The MrpA model shows the N-terminal domain sharing a central core with NuoL, composed of 14 TMS, and a C-terminal extension forming a piston, composed of a predicted TMS and a long helix terminating with a TMS similar to that of NuoL. The piston sequence is 50 residues shorter than the NuoL piston, which brings the N-terminal and C-terminal domains of MrpA closer. The model suggests that MrpA has the potential to accommodate a single subunit of the Mrp complex, in contrast to complex I from E. coli, where the longer piston accommodates two antiporter-like subunits (21). The MrpA model has been shown to accommodate the antiporter-like NuoN subunit of complex I, which is homologous to MrpD (21). A role for the MrpA_C domain is consistent with an essential role for the domain in the antiport activity of the Bacillus pseudofirmus Mrp complex (43).
FIG 5.
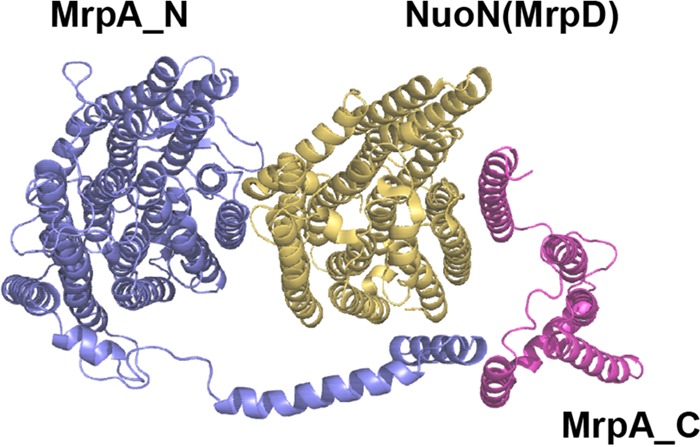
M. acetivorans MrpA modeled with NuoN. The MrpA N-terminal and C-terminal domains are shown in purple and magenta, respectively. See Materials and Methods for the modeling procedures.
Proposed mechanism.
The model of MrpA provides a basis for proposing an ion translocation mechanism for the N-terminal domain of MrpA (Fig. 6). The model features two half-closed symmetry-related channels that are linked, forming a continuous path from the cytoplasm to the periplasm, analogous to NuoL (21). The periplasm in M. acetivorans is defined as the space between the cytoplasmic membrane and the cell wall. The cytoplasm-facing channel 1 is near TM5 and TM7, and the periplasm-facing channel 2 is near TM12. Key residues in NuoL channel 1 (K229 and E144) and channel 2 (K399) correspond to conserved residues K227, E141, and K402 in MrpA (see Fig. S4 in the supplemental material). Indeed, an important role for K227 and E141 in antiport activity is supported by site-directed replacement analyses of cognate residues in MrpA homologs from the domain Bacteria (5). The essential D400 in NuoL channel 2 is replaced with E403 in MrpA (37). Similar to NuoL, the MrpA model displays cavities facing the cytoplasmic and periplasmic sides of the membrane. The cytoplasm-facing cavity is formed by R112, E141, S147, T171, K227, and T255. The periplasm-facing cavity contains residues S309, S312, Y316, H339, S401, K402, E403, T439, Y442, and S443. The two cavities are connected by charged and polar residues (H252, K347, H343, T310, and S253) conserved in NuoL that complete the ion translocation pathway. The invariant residue H254, which links the two half channels in NuoL, corresponds to H252 of MrpA TM8. The motif 251-LHSATM-256 in TM8 of MrpA is largely conserved in NuoL (see Fig. S4 in the supplemental material), consistent with a role common to both complexes. TM7 and TM12 in MrpA are interrupted in the middle of the membrane by extended loops containing P232 and P394 that are conserved in the cognate loops of NuoL (see Fig. S4 in the supplemental material). Both helices are probably involved in ion transport by introducing flexibility to the structure, allowing conformational changes driven by the piston-like structure that promote ion translocation, as proposed for NuoL (21).
FIG 6.
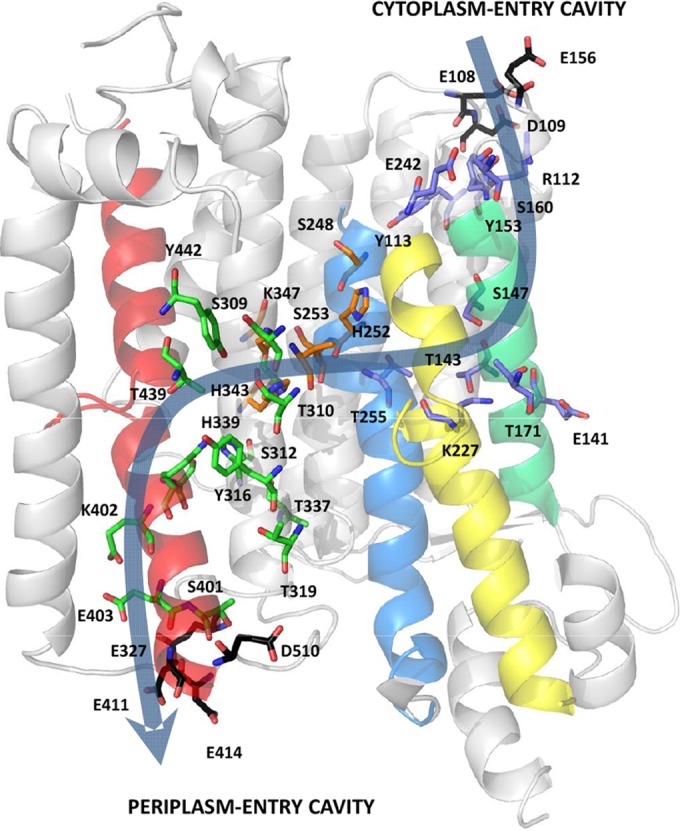
Proposed ion translocation channel in MrpA from M. acetivorans. Shown in stick representation are polar and charged residues forming the transmembrane channel, composed of the cytoplasm-facing cavity (blue), the periplasm-facing cavity (green), and a zone connecting the cavities (orange). Negatively charged residues that form cytoplasm- and periplasm-facing patches at TM5 and TM12, respectively, are shown in black. Main TMS are colored as follows: TMS5 (green), TMS7 (yellow), TMS8 (blue), and TMS12 (red). The transmembrane ion translocation channel is shown by a light-blue arrow. The figure was generated with the PyMOL Molecular Graphics System version 1.2r1 (Schrödinger LLC).
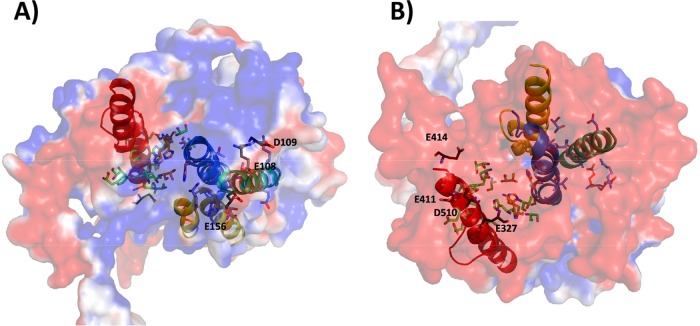
The surface electrostatic potential of the N-terminal domain of MrpA from M. acetivorans shows a negatively charged patch formed by D109, E108, and E156 in TM4-TM5 (Fig. 7). This patch is a potential interface with the cytoplasm, as it corresponds to the entry of the cytoplasm-facing cavity of NuoL (21). Since MrpA was modeled with the NuoL template in the closed conformation, the proposed Mrp interface is also closed. The MrpA model displays a second negatively charged patch on the surface formed by residues E360, E375, E454, D458, and D460 in TM12. This patch is larger than that in TM4-TM5, and the possibility that it interfaces with the cytoplasm cannot be excluded. The surface electrostatic potential, viewed from the periplasm-facing perspective, highlights a strong negatively charged character defined by residues E327, E411, E414, and D510, with the potential to participate in ion translocation.
FIG 7.
M. acetivorans MrpA N-terminal domain electrostatic surface potentials. (A) Cytoplasm-facing surface. (B) Periplasm-facing surface. The black residues represent negatively charged patches on the TM5 and TM12 surfaces. Residues and TM-forming cavities are represented as in Fig. 6. The surface potential was calculated using the PDB2PQR server (http://nbcr-222.ucsd.edu/pdb2pqr_2.0.0/) (57). The figure was generated with the PyMOL Molecular Graphics System version 1.2r1 (Schrödinger LLC).
The modeled structure provides a framework for proposing a role for MrpA in the Na+/H+ antiporter activity of antiporter-deficient E. coli strains expressing MrpA from M. acetivorans. Conceivably, NuoN subunits from E. coli complex I combine with MrpA to produce a functional Na+/H+ antiporter unit, as shown in Fig. 5. This proposal is consistent with the ability of NuoL and NuoN to replace MrpA and MrpD, respectively, of the Mrp complex in Bacillus species in which the subunits are homologous to MrpA and MrpD of M. acetivorans (25). Thus, we propose that MrpA functions together with MrpD to form an MrpAD subcomplex in M. acetivorans catalyzing Na+/H+ antiport independently of MrpBCEFG. The piston of MrpA is postulated to stabilize the subcomplex, analogous to what has been proposed for the piston in complex I (34, 44). The proposed MrpAD subcomplex is consistent with the finding that an MrpA/MrpD homolog encoded by a single gene from N. thermophilus catalyzes Na+/H+ antiport in E. coli KNabc. The proposed MrpAD subcomplex is also consistent with the hypothesis that the homologous MrpA and MrpD subunits of B. subtilis translocate Na+ and H+, respectively, in opposite directions (25). Ion translocations in MrpA and MrpD are likely to be driven by conformational coupling, analogous to the proposed conformation-driven translocation of protons for NuoM and NuoN in complex I (21). It is tempting to speculate that an MrpBCEFG subcomplex of M. acetivorans samples the scale of Na+ and H+ gradients and induces conformational changes in the MrpAD subcomplex that modulate antiporter activity, thereby adjusting the Na+/H+ ratio optimally for ATP synthase.
On the other hand, Na+/H+ antiport catalyzed by MrpA alone cannot be ruled out at this juncture. It has been reported that the H+-pumping NuoL subunit of complex I can replace the postulated Na+-pumping MrpA subunit of the MrpABCDEFG complex from B. subtilis, consistent with the ability of NuoL and MrpA to translocate both Na+ and H+ (25). MrpA may form a homodimer in head-to-tail fashion, with the N-terminal domain interacting with the piston domain. The ionizable D563 in the piston of E. coli NuoL is proposed to be involved in proton translocation (39). Thus, residues D542, D553, and D565 in the piston region of MrpA from M. acetivorans may be involved in antiporter activity.
Conclusions.
Characterization of the MrpA subunit in the MrpABCDEFG complex from M. acetivorans of the domain Archaea has shown that MrpA plays a role in Na+/H+ antiport activity, and that activity is independent of an intact MrpABCDEFG complex. A mechanism was proposed in which an MrpAD subcomplex catalyzes the activity modulated by an MrpBCEFG subcomplex. Shared properties support the proposal that complex I evolved by recruitment of subunits from MrpA complexes of ancient origin. Finally, bioinformatics analyses indicate that Mrp complexes function in diverse methanogenic pathways.
MATERIALS AND METHODS
Materials.
Antibiotics (chloramphenicol, kanamycin, and ampicillin), NADH, RNase A and DNase I (both from bovine pancreas), acridine orange probe, and Bis-Tris propane were purchased from Sigma (St. Louis, MO). Protein precipitation and DNA hydration solutions were from Qiagen (Maryland, USA). Restriction enzymes NdeI and SacII were from New England BioLabs. The pET21a(+) cloning vector was from Invitrogen, E. coli Nova Blue Singles competent cells were purchased from Novagen. All the salts were analytical grade and were purchased from Sigma.
DNA isolation and cloning of mrpA.
Total DNA isolation was carried out using a 1.5-ml aliquot of methanol-grown M. acetivorans cells in the early stationary phase. Cells were harvested by centrifugation at 16,000 × g for 2 min, and the pellet was resuspended in 0.5 ml of Tris-EDTA (TE) buffer at pH 7.0. Then, 0.015 ml 10% (wt/vol) SDS and 1 IU of RNase were added to the sample, and the tube was gently mixed. The sample was further incubated at 37°C for 60 min. Then, the protein precipitation solution was added, and the tube was strongly vortexed and further centrifuged at 16,162 × g for 10 min. The supernatant solution was mixed with absolute 2-propanol and centrifuged again. The pellet containing DNA was incubated at −20°C for 60 min. The DNA was air dried, and 70% (vol/vol) ethanol was added to wash the DNA pellet, which was air dried again. Finally, 0.05 ml of the DNA hydration solution was added to suspend the DNA. Approximately 500 μg DNA/ml was typically obtained with this protocol.
The cloning of mrpA from M. acetivorans was accomplished by PCR using primers designed based on the sequence of the MA4572 locus in the operon encoding the seven MrpABCDEFG subunits of the complex (30). The primers used were forward, 5′-GGCCGGCATATGGATCCATTTAATGCA-3′, and reverse, 5′-TTATTCCTTACCTCCCTTCAC-3′. The primers contained sites for the restriction enzymes NdeI and SacII, respectively. The PCR product was inserted into the pET21a(+) cloning vector and used to transform E. coli NovaBlue Singles competent cells. Vector pET21a(+) containing the insert (designated pET-MAmrpA) was isolated from the NovaBlue cells and, after confirming the sequence of mrpA, was used to transform the sodium-sensitive E. coli strains EP432 and KNabc. Both E. coli strains lack genes encoding the two Na+/H+ antiporters, NhaA and NhaB, whereas strain KNabc also lacks chaA, which encodes one of the Na+ extrusion systems important for growth at high pH. Clones containing pET-MAmrpA were selected after screening for growth on plates containing LB medium supplemented with 100 μg ampicillin/ml (see Fig. S1 in the supplemental material). The selected clones were designated mrpA EP432 and mrpA KNabc, respectively.
Culture conditions and strains.
M. acetivorans wild-type strain C2A was cultured at 37°C in high-salt medium with 100 mM methanol as the growth substrate, as previously described (45). The Na+/H+ antiporter-deficient E. coli strains EP432 (melB [Li+ dependent] ΔnhaA1::kan ΔnhaB1::cam ΔlacZY thr-1) and KNabc (TG1 [ΔnhaA ΔnhaB ΔchaA]) are described elsewhere (46, 47). Both strains were maintained in LBK medium, obtained by replacing NaCl in LB medium with KCl (10 g/liter) (48). The parental E. coli strain K-12 and the mutant E. coli strain EP432 were grown without antibiotics in LB and LBK media, respectively, whereas LBK medium for growth of E. coli strain KNabc was supplemented with 30 μg kanamycin/ml and 17.5 μg chloramphenicol/ml. The medium for growth of E. coli strains mrpA EP432 and mrpA KNabc was regular LB medium further supplemented with 50 μg ampicillin/ml. When the resistance to sodium was tested, NaCl in LB medium was omitted and the desired NaCl concentration was adjusted using a 5 M NaCl sterile stock solution. When different pH values were required, cells were grown in LBK medium, and 1 M KOH sterile stock solution was used to adjust the pH. The cultures (5 ml) were grown overnight (10 to 11 h) at 37°C with orbital shaking (200 rpm), and the final optical density at 600 nm (OD600) was recorded.
Preparation of everted membrane vesicles and measurement of Na+/H+ antiporter activity.
Strains of E. coli were cultured in 1 liter of LB medium containing KCl (135 mM final concentration) for growth of E. coli strains EP432 and KNabc or NaCl (171 mM final concentration) for growth of strains mrpA EP432 and mrpA KNabc and the parental strain K-12. The medium was also supplemented with antibiotics appropriate for each strain, as listed above. When culturing E. coli strains mrpA EP432 and mrpA KNabc, it was necessary to add 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at the beginning to induce MrpA synthesis, which was required for growth in the presence of the regular LB medium containing 171 mM NaCl (10 g/liter). All the cultures were grown overnight (11 to 12 h) at 37°C with 200-rpm orbital shaking. Preparation of membrane vesicles was carried out similarly to that described previously for M. acetivorans (49).
The assay of ΔpH-dependent antiport activity was performed as follows. Everted vesicles (50 to 75 μg protein) were incubated in 2 ml of 50 mM Bis-Tris propane buffer containing 140 mM choline-Cl, 10% (vol/vol) glycerol, 5 mM MgSO4, and 0.5 μM acridine orange adjusted to the indicated pH with 1 M HCl. The ΔpH (acid inside) was generated by addition of 0.5 mM NADH or 5 mM d-lactate. The acridine orange was excited at 492 nm, and the emission was recorded at 523 nm (12). Formation of ΔpH (quenching of fluorescence) and collapse of the gradient (dequenching of fluorescence) were measured after the addition of NaCl, LiCl, or KCl. Maximum dequenching was determined by adding 5 μM CCCP to dissipate the remaining ΔpH and return the fluorescence to baseline. Antiport activity was determined by the percentage of maximum dequenching after the signal reached equilibrium. The apparent Km was estimated from the concentration of cation that produced half-maximal dequenching prior to the addition of CCCP. Although not based on rates, the method was shown to be a good estimate of the Km, as described previously (4).
Molecular modeling.
To gain an indication of the structure of MrpA from M. acetivorans, a molecular model was built by merging independent models of the MrpA_N and MrpA_C domains of MrpA. The MrpA_N model was generated using a template based on the homologous NuoL subunit of E. coli complex I (Protein Data Bank [PDB] accession no. 3RKO), of which the crystal structure is known (21). The MrpA_C model was generated using a template based on the homologous NuoJ subunits of E. coli complex I (PDB accession no. 3RKO) and Thermus thermophilus complex I (PDB accession no. 4HEA), of which the crystal structures are also known (35). Preliminary models of MrpA_N and MrpA_C were built using the I-TASSER server (50) and were then used to construct high-quality three-dimensional homology models using MODELLER 9v12 (https://salilab.org/modeller/download_installation.html) (51). The models were verified with PROCHECK v3.5 (see Table S1 and Fig. S2 and S3 in the supplemental material), which qualifies combinations of the ϕ and ψ angles of residues in allowed zones of the Ramachandran plot (52). The analysis indicated that the models have high enough confidence to perform structural analyses.
A tentative model of MrpA from M. acetivorans was constructed with the MrpA_N and MrA_C models using the PyMOL Molecular Graphics System version 1.2r1 (Schrödinger, LLC) with a template derived from the NuoLNMJ subunits of E. coli complex I (3RKO). The MrpA_N model was aligned to the NuoL position of the template, followed by alignment of NuoN to the position occupied by NuoM. The MrpA_C model was aligned to the position occupied by NuoJ and adjusted to reside closer to NuoN.
Supplementary Material
ACKNOWLEDGMENTS
The work was supported by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences, of the U.S. Department of Energy through grant no. DE-FG02-95ER20198 MOD16 (J.G.F.) and National Science Foundation grant no. 0820734 (J.G.F.). The work was also supported by grant no. 156969 from CONACyT, Mexico (R.J.-C.), and grants 979/2016 and 169/2016 from DAIP-Universidad de Guanajuato, Mexico (C.D.-P.).
We acknowledge Prashanti Iyer for assistance in constructing the phylogenetic tree. E. coli strains EP432 (melB [Li+ dependent] ΔnhaA1::kan ΔnhaB1::cam ΔlacZY thr-1) and KNabc (TG1 [ΔnhaA ΔnhaB ΔchaA]) were kindly provided by Etana Padan and Terry Krulwich.
R.J.-C., C.D.-P., J.S.R.-Z., and J.G.F. conceived the experiments; R.J.-C., C.D.-P., and J.S.R.-Z. performed the experiments; and R.J.-C., C.D.-P., J.S.R.-Z., and J.G.F. interpreted the results and contributed to writing the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00662-16.
REFERENCES
- 1.Swartz TH, Ikewada S, Ishikawa O, Ito M, Krulwich TA. 2005. The Mrp system: a giant among monovalent cation/proton antiporters? Extremophiles 9:345–354. doi: 10.1007/s00792-005-0451-6. [DOI] [PubMed] [Google Scholar]
- 2.Morino M, Suzuki T, Ito M, Krulwich TA. 2014. Purification and functional reconstitution of a seven-subunit Mrp-type Na+/H+ antiporter. J Bacteriol 196:28–35. doi: 10.1128/JB.01029-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kajiyama Y, Otagiri M, Sekiguchi J, Kosono S, Kudo T. 2007. Complex formation by the mrpABCDEFG gene products, which constitute a principal Na+/H+ antiporter in Bacillus subtilis. J Bacteriol 189:7511–7514. doi: 10.1128/JB.00968-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swartz TH, Ito M, Ohira T, Natsui S, Hicks DB, Krulwich TA. 2007. Catalytic properties of Staphylococcus aureus and Bacillus members of the secondary cation/proton antiporter-3 (Mrp) family are revealed by an optimized assay in an Escherichia coli host. J Bacteriol 189:3081–3090. doi: 10.1128/JB.00021-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morino M, Natsui S, Ono T, Swartz TH, Krulwich TA, Ito M. 2010. Single site mutations in the hetero-oligomeric Mrp antiporter from alkaliphilic Bacillus pseudofirmus OF4 that affect Na+/H+ antiport activity, sodium exclusion, individual Mrp protein levels, or Mrp complex formation. J Biol Chem 285:30942–30950. doi: 10.1074/jbc.M110.118661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito M, Guffanti AA, Krulwich TA. 2001. Mrp-dependent Na+/H+ antiporters of Bacillus exhibit characteristics that are unanticipated for completely secondary active transporters. FEBS Lett 496:117–120. doi: 10.1016/S0014-5793(01)02417-6. [DOI] [PubMed] [Google Scholar]
- 7.Ito M, Guffanti AA, Oudega B, Krulwich TA. 1999. mrp, a multigene, multifunctional locus in Bacillus subtilis with roles in resistance to cholate and to Na+ and in pH homeostasis. J Bacteriol 181:2394–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kashyap DR, Botero LM, Lehr C, Hassett DJ, McDermott TR. 2006. A Na+:H+ antiporter and a molybdate transporter are essential for arsenite oxidation in Agrobacterium tumefaciens. J Bacteriol 188:1577–1584. doi: 10.1128/JB.188.4.1577-1584.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Putnoky P, Kereszt A, Nakamura T, Endre G, Grosskopf E, Kiss P, Kondorosi A. 1998. The pha gene cluster of Rhizobium meliloti involved in pH adaptation and symbiosis encodes a novel type of K+ efflux system. Mol Microbiol 28:1091–1101. doi: 10.1046/j.1365-2958.1998.00868.x. [DOI] [PubMed] [Google Scholar]
- 10.Blanco-Rivero A, Leganes F, Fernandez-Valiente E, Calle P, Fernandez-Pinas F. 2005. mrpA, a gene with roles in resistance to Na+ and adaptation to alkaline pH in the cyanobacterium Anabaena sp. PCC7120. Microbiology 151:1671–1682. doi: 10.1099/mic.0.27848-0. [DOI] [PubMed] [Google Scholar]
- 11.Ito M, Guffanti AA, Wang W, Krulwich TA. 2000. Effects of nonpolar mutations in each of the seven Bacillus subtilis mrp genes suggest complex interactions among the gene products in support of Na+ and alkali but not cholate resistance. J Bacteriol 182:5663–5670. doi: 10.1128/JB.182.20.5663-5670.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dzioba-Winogrodzki J, Winogrodzki O, Krulwich TA, Boin MA, Hase CC, Dibrov P. 2009. The Vibrio cholerae Mrp system: cation/proton antiport properties and enhancement of bile salt resistance in a heterologous host. J Mol Microbiol Biotechnol 16:176–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kosono S, Haga K, Tomizawa R, Kajiyama Y, Hatano K, Takeda S, Wakai Y, Hino M, Kudo T. 2005. Characterization of a multigene-encoded sodium/hydrogen antiporter (Sha) from Pseudomonas aeruginosa: its involvement in pathogenesis. J Bacteriol 187:5242–5248. doi: 10.1128/JB.187.15.5242-5248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanco-Rivero A, Leganes F, Fernandez-Valiente E, Fernandez-Pinas F. 2009. mrpA (all1838), a gene involved in alkali and Na+ sensitivity, may also have a role in energy metabolism in the cyanobacterium Anabaena sp. strain PCC 7120. J Plant Physiol 166:1488–1496. doi: 10.1016/j.jplph.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Jasso-Chavez R, Apolinario EE, Sowers KR, Ferry JG. 2013. MrpA functions in energy conversion during acetate-dependent growth of Methanosarcina acetivorans. J Bacteriol 195:3987–3994. doi: 10.1128/JB.00581-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schut GJ, Boyd ES, Peters JW, Adams MW. 2013. The modular respiratory complexes involved in hydrogen and sulfur metabolism by heterotrophic hyperthermophilic archaea and their evolutionary implications. FEMS Microbiol Rev 37:182–203. doi: 10.1111/j.1574-6976.2012.00346.x. [DOI] [PubMed] [Google Scholar]
- 17.Boyd ES, Schut GJ, Adams MWW, Peters JW. 2014. Hydrogen metabolism and the evolution of biological respiration. Microbe 9:361–367. [Google Scholar]
- 18.Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. 2013. Crystal structure of the entire respiratory complex I. Nature 494:443–448. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathiesen C, Hagerhall C. 2002. Transmembrane topology of the NuoL, M and N subunits of NADH:quinone oxidoreductase and their homologues among membrane-bound hydrogenases and bona fide antiporters. Biochim Biophys Acta 1556:121–132. doi: 10.1016/S0005-2728(02)00343-2. [DOI] [PubMed] [Google Scholar]
- 20.Moparthi VK, Kumar B, Al-Eryani Y, Sperling E, Gorecki K, Drakenberg T, Hagerhall C. 2014. Functional role of the MrpA- and MrpD-homologous protein subunits in enzyme complexes evolutionary related to respiratory chain complex I. Biochim Biophys Acta 1837:178–185. doi: 10.1016/j.bbabio.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 21.Efremov RG, Sazanov LA. 2011. Structure of the membrane domain of respiratory complex I. Nature 476:414–420. doi: 10.1038/nature10330. [DOI] [PubMed] [Google Scholar]
- 22.Mathiesen C, Hagerhall C. 2003. The ‘antiporter module’ of respiratory chain complex I includes the MrpC/NuoK subunit—a revision of the modular evolution scheme. FEBS Lett 549:7–13. doi: 10.1016/S0014-5793(03)00767-1. [DOI] [PubMed] [Google Scholar]
- 23.Sousa FL, Nelson-Sathi S, Martin WF. 2016. One step beyond a ribosome: the ancient anaerobic core. Biochim Biophys Acta 1857:1027–1038. doi: 10.1016/j.bbabio.2016.04.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kajiyama Y, Otagiri M, Sekiguchi J, Kudo T, Kosono S. 2009. The MrpA, MrpB, and MrpD subunits of the Mrp antiporter complex in Bacillus subtilis contain membrane-embedded and essential acidic residues. Microbiology 155:2137–2147. doi: 10.1099/mic.0.025205-0. [DOI] [PubMed] [Google Scholar]
- 25.Moparthi VK, Kumar B, Mathiesen C, Hagerhall C. 2011. Homologous protein subunits from Escherichia coli NADH:quinone oxidoreductase can functionally replace MrpA and MrpD in Bacillus subtilis. Biochim Biophys Acta 1807:427–436. doi: 10.1016/j.bbabio.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Morino M, Natsui S, Swartz TH, Krulwich TA, Ito M. 2008. Effect of single gene deletions of mrpA-G and mrpE point mutations on activity of the Mrp Na+/H+ antiporter of alkaliphilic Bacillus and formation of hetero-oligomeric Mrp complex. Biochim Biophys Acta 1777:S26–S27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morino M, Natsui S, Swartz TH, Krulwich TA, Ito M. 2008. Single gene deletions of mrpA to mrpG and mrpE point mutations affect activity of the Mrp Na+/H+ antiporter of alkaliphilic Bacillus and formation of hetero-oligomeric Mrp complexes. J Bacteriol 190:4162–4172. doi: 10.1128/JB.00294-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mesbah NM, Cook GM, Wiegel J. 2009. The halophilic alkalithermophile Natranaerobius thermophilus adapts to multiple environmental extremes using a large repertoire of Na+(K+)/H+ antiporters. Mol Microbiol 74:270–281. doi: 10.1111/j.1365-2958.2009.06845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlegel K, Leone V, Faraldo-Gomez JD, Muller V. 2012. Promiscuous archaeal ATP synthase concurrently coupled to Na+ and H+ translocation. Proc Natl Acad Sci U S A 109:947–952. doi: 10.1073/pnas.1115796109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Q, Li L, Rejtar T, Lessner DJ, Karger BL, Ferry JG. 2006. Electron transport in the pathway of acetate conversion to methane in the marine archaeon Methanosarcina acetivorans. J Bacteriol 188:702–710. doi: 10.1128/JB.188.2.702-710.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlegel K, Welte C, Deppenmeier U, Muller V. 2012. Electron transport during aceticlastic methanogenesis by Methanosarcina acetivorans involves a sodium-translocating Rnf complex. FEBS J 279:4444–4452. doi: 10.1111/febs.12031. [DOI] [PubMed] [Google Scholar]
- 32.Welte C, Deppenmeier U. 2011. Proton translocation in methanogens. Methods Enzymol 494:257–280. doi: 10.1016/B978-0-12-385112-3.00013-5. [DOI] [PubMed] [Google Scholar]
- 33.Williams TJ, Burg DW, Ertan H, Raftery MJ, Poljak A, Guilhaus M, Cavicchioli R. 2010. Global proteomic analysis of the insoluble, soluble, and supernatant fractions of the psychrophilic archaeon Methanococcoides burtonii. Part II: the effect of different methylated growth substrates. J Proteome Res 9:653–663. doi: 10.1021/pr9005102. [DOI] [PubMed] [Google Scholar]
- 34.Virzintiene E, Moparthi VK, Al-Eryani Y, Shumbe L, Gorecki K, Hagerhall C. 2013. Structure and function of the C-terminal domain of MrpA in the Bacillus subtilis Mrp-antiporter complex: the evolutionary progenitor of the long horizontal helix in complex I. FEBS Lett 587:3341–3347. doi: 10.1016/j.febslet.2013.08.027. [DOI] [PubMed] [Google Scholar]
- 35.Efremov RG, Baradaran R, Sazanov LA. 2010. The architecture of respiratory complex I. Nature 465:441–445. doi: 10.1038/nature09066. [DOI] [PubMed] [Google Scholar]
- 36.Kosono S, Kajiyama Y, Kawasaki S, Yoshinaka T, Haga K, Kudo T. 2006. Functional involvement of membrane-embedded and conserved acidic residues in the ShaA subunit of the multigene-encoded Na+/H+ antiporter in Bacillus subtilis. Biochim Biophys Acta 1758:627–635. doi: 10.1016/j.bbamem.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 37.Nakamaru-Ogiso E, Kao MC, Chen H, Sinha SC, Yagi T, Ohnishi T. 2010. The membrane subunit NuoL(ND5) is involved in the indirect proton pumping mechanism of Escherichia coli complex I. J Biol Chem 285:39070–39078. doi: 10.1074/jbc.M110.157826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vinothkumar KR, Zhu J, Hirst J. 2014. Architecture of mammalian respiratory complex I. Nature 515:80–84. doi: 10.1038/nature13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steimle S, Willistein M, Hegger P, Janoschke M, Erhardt H, Friedrich T. 2012. Asp563 of the horizontal helix of subunit NuoL is involved in proton translocation by the respiratory complex I. FEBS Lett 586:699–704. doi: 10.1016/j.febslet.2012.01.056. [DOI] [PubMed] [Google Scholar]
- 40.Rothman DH, Fournier GP, French KL, Alm EJ, Boyle EA, Cao C, Summons RE. 2014. Methanogenic burst in the end-Permian carbon cycle. Proc Natl Acad Sci U S A 111:5462–5467. doi: 10.1073/pnas.1318106111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deppenmeier U, Johann A, Hartsch T, Merkl R, Schmitz RA, Martinez-Arias R, Henne A, Wiezer A, Baumer S, Jacobi C, Bruggemann H, Lienard T, Christmann A, Bomeke M, Steckel S, Bhattacharyya A, Lykidis A, Overbeek R, Klenk HP, Gunsalus RP, Fritz HJ, Gottschalk G. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between Bacteria and Archaea. J Mol Microbiol Biotechnol 4:453–461. [PubMed] [Google Scholar]
- 42.Allen MA, Lauro FM, Williams TJ, Burg D, Siddiqui KS, De Francisci D, Chong KW, Pilak O, Chew HH, De Maere MZ, Ting L, Katrib M, Ng C, Sowers KR, Galperin MY, Anderson IJ, Ivanova N, Dalin E, Martinez M, Lapidus A, Hauser L, Land M, Thomas T, Cavicchioli R. 2009. The genome sequence of the psychrophilic archaeon, Methanococcoides burtonii: the role of genome evolution in cold adaptation. ISME J 3:1012–1035. doi: 10.1038/ismej.2009.45. [DOI] [PubMed] [Google Scholar]
- 43.Morino M, Ogoda S, Krulwich TA, Ito M. Differences in the phenotypic effects of mutations in homologous MrpA and MrpD subunits of the multi-subunit Mrp-type Na+/H+ antiporter. Extremophiles, in press. [DOI] [PubMed] [Google Scholar]
- 44.Torres-Bacete J, Sinha PK, Matsuno-Yagi A, Yagi T. 2011. Structural contribution of C-terminal segments of NuoL (ND5) and NuoM (ND4) subunits of complex I from Escherichia coli. J Biol Chem 286:34007–34014. doi: 10.1074/jbc.M111.260968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sowers KR, Baron SF, Ferry JG. 1984. Methanosarcina acetivorans sp. nov., an acetotrophic methane-producing bacterium isolated from marine sediments. Appl Environ Microbiol 47:971–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pinner E, Kotler Y, Padan E, Schuldiner S. 1993. Physiological role of nhaB, a specific Na+/H+ antiporter in Escherichia coli. J Biol Chem 268:1729–1734. [PubMed] [Google Scholar]
- 47.Nozaki K, Inaba K, Kuroda T, Tsuda M, Tsuchiya T. 1996. Cloning and sequencing of the gene for Na+/H+ antiporter of Vibrio parahaemolyticus. Biochem Biophys Res Commun 222:774–779. doi: 10.1006/bbrc.1996.0820. [DOI] [PubMed] [Google Scholar]
- 48.Herz K, Vimont S, Padan E, Berche P. 2003. Roles of NhaA, NhaB, and NhaD Na+/H+ antiporters in survival of Vibrio cholerae in a saline environment. J Bacteriol 185:1236–1244. doi: 10.1128/JB.185.4.1236-1244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suharti S, Wang M, de Vries S, Ferry JG. 2014. Characterization of the RnfB and RnfG subunits of the Rnf complex from the archaeon Methanosarcina acetivorans. PLoS One 9:e97966. doi: 10.1371/journal.pone.0097966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sali A, Blundell TL. 1993. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 52.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 53.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 54.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 55.Zuckerkandl E, Pauling L. 1965. Evolutionary divergence and convergence in proteins, p 97–166. In Bryson V, Vogel HJ (ed), Evolving genes and proteins. Academic Press, New York. [Google Scholar]
- 56.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. 2004. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res 32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.