Table 2.
Shown in (a) are the estimates of the log hazard ratio (logHR) along with their corresponding standard errors (SE) and p-values by fitting stratified Cox model integrating information from 4 independent studies with imputed miRNA based on the SMC method and the nuclear norm minimization (NNM); and Cox model to the TCGA test data with original observed miRNA (Ori.). Shown also are the estimates for each individual studies by fitting separate Cox models with imputed miRNA.
| (a) Integrated Analysis with
Imputed miRNA vs Single study with observed miRNA | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| logHR | SE | p-value | ||||||||
| Ori. | SMC | NNM | Ori. | SMC | NNM | Ori. | SMC | NNM | ||
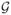
|
.067 | .143 | .168 | .041 | .034 | .028 | .104 | .000 | .000 | |
|
|
−.012 | −.019 | −.013 | .009 | .006 | .012 | .218 | .001 | .283 | |
|
|
.023 | .018 | −.005 | .014 | .009 | .014 | .092 | .039 | .725 | |
| (b) Estimates for Individual
Studies with Imputed miRNA from the SMC method | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| logHR | SE | p-value | |||||||||||
| TCGA | TOTH | DRES | BONO | TCGA | TOTH | DRES | BONO | TCGA | TOTH | DRES | BONO | ||
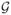
|
.051 | .377 | .174 | .311 | .048 | .069 | .132 | .117 | .286 | .000 | .187 | .008 | |
|
|
−.014 | −.021 | −.031 | −.010 | .011 | .012 | .014 | .014 | .207 | .082 | .030 | .484 | |
|
|
.014 | .045 | −.021 | .036 | .016 | .018 | .022 | .019 | .391 | .009 | .336 | .054 | |
| (c) Estimates for Individual
Studies with Imputed miRNA from the NNM method | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| logHR | SE | p-value | |||||||||||
| TCGA | TOTH | DRES | BONO | TCGA | TOTH | DRES | BONO | TCGA | TOTH | DRES | BONO | ||
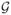
|
.082 | .405 | .361 | .258 | .037 | .066 | .114 | .088 | .028 | .000 | .002 | .003 | |
|
|
−.045 | .016 | .055 | −.008 | .021 | .026 | .031 | .023 | .034 | .544 | .076 | .721 | |
|
|
.008 | −.086 | −.043 | .019 | .026 | .027 | .034 | .029 | .758 | .002 | .201 | .496 | |
