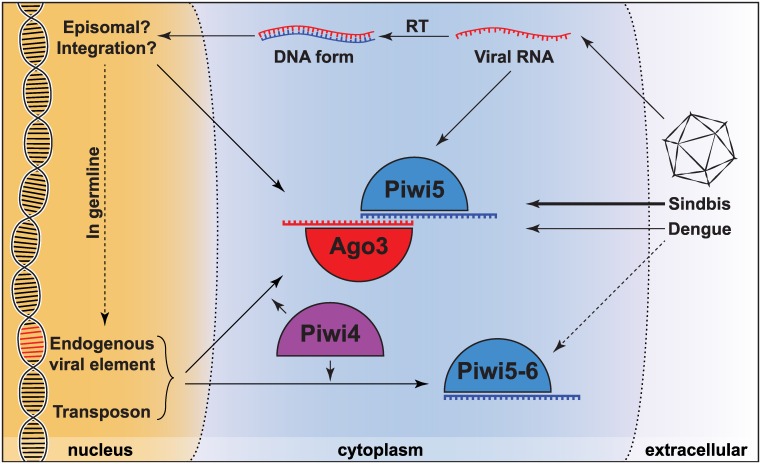
Fig 3. Model for piRNA biogenesis in Aedes aegypti.
RNA molecules from varying sources are processed differently by the piRNA machinery in Ae. aegypti. Upon acute infection, Sindbis virus RNA is processed into ping-pong–dependent piRNAs involving PIWI proteins Piwi5 and Ago3. In contrast, dengue virus RNA can also be processed into piRNAs by Piwi6. Transposon-derived piRNAs associate primarily with Piwi5 and Piwi6; however, some transposon RNAs feed into the ping-pong loop and give rise to Ago3-bound secondary piRNAs. Additionally, the production of transposon piRNAs is dependent on Piwi4 in an indirect manner, as transposon-derived piRNAs are not loaded in Piwi4, but knockdown of Piwi4 does reduce their numbers. Viral RNA may directly enter the piRNA machinery; additionally, viral RNA is reverse transcribed to produce a DNA form of the virus (vDNA). The vDNA may either remain episomal or integrate into the host genome. Putative vDNA-derived transcripts may serve as additional precursors for vpiRNA production. Moreover, when genome integration occurs in the germline, the vDNA fragment forms a novel endogenous viral element (EVE) that may lead to the production of EVE-derived piRNAs.