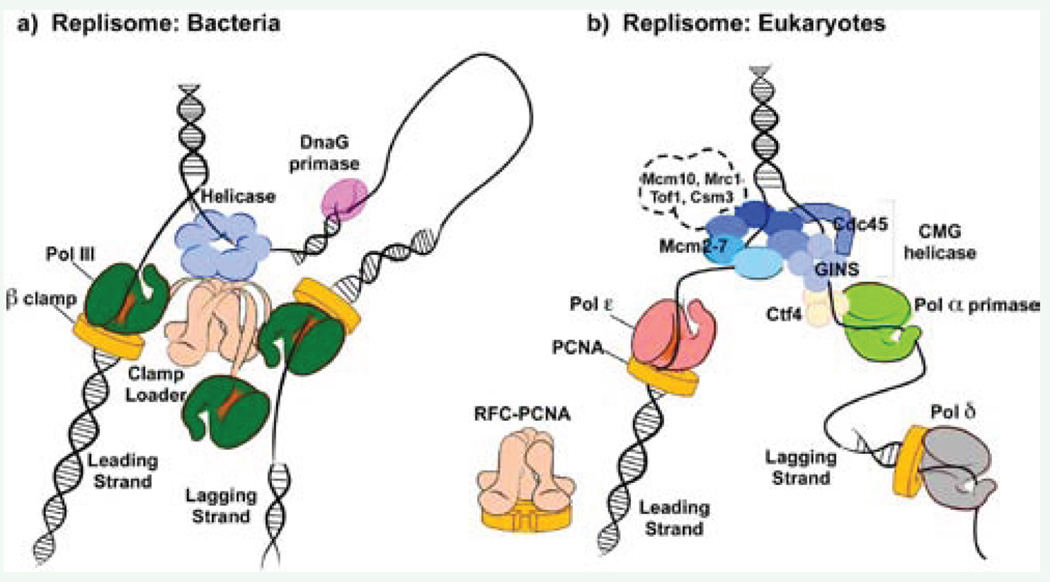
Figure 3.
Comparison of replisomes from bacteria and eukaryotes. a) The bacterial replisome is organized by a central clamp loading machine (beige) that has C-terminal extension, which extrude from the top and bind the hexameric helicase (blue) that encircles the lagging strand. The clamp loader extensions also bind three copies of Pol III (green), the replicative DNA polymerases. One Pol III functions on the continuous leading strand, tethered to DNA by the beta clamp (yellow). The other two Pol III molecules extend lagging strand fragments that are initiated by short RNA primers synthesized by primase (pink). b) The eukaryotic replisome is organized by the CMG helicase (blue), which functions with Pol epsilon (red) on the leading strand and bind Pol alpha-primase (green) through a Ctf4 trimer (light yellow) on the lagging strand. Lagging strand primers are extended by Pol delta (grey). Both Pol epsilon and Pol delta function with a PCNA clamp (yellow). Whether the RFC clamp loader and Pol delta directly connect to other replisome proteins is not known. Other factors that move with replisomes are shown within the dashed outline; their exact connection points are not yet known.