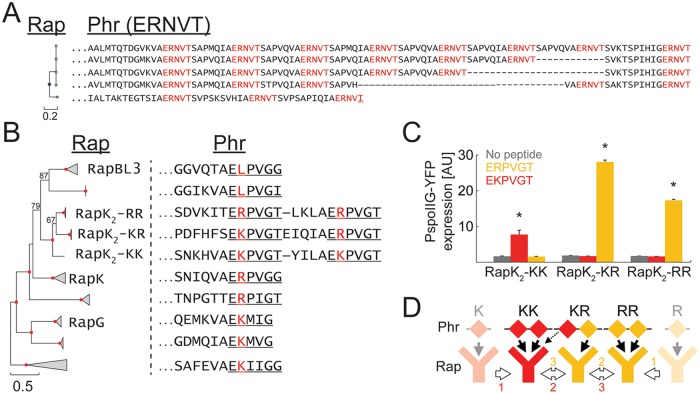
Fig 4. Phr sequence duplication and neofunctionalization.
(A) Phylogenetic tree of five specific Rap variants (left) and their respective Phr sequence. The first twenty amino acids of the Phr sequence are omitted. Marked in red are repeated sequences of the putative Phr autoinducer. Underlined in the bottom sequence is a putative autoinducer sequence that deviates from the consensus autoinducer. (B) Phylogenetic tree of 140 Rap variants related to RapK. Triangles mark a cluster with the same putative autoinducer. Triangle lengths mark the mean distance between the ancestral node of the cluster to specific leaves. Red squares mark nodes with 100% support (out of 100 bootstrap repeats). Several additional nodes are marked by the number of bootstrap repeats supporting them. If a variant from a cluster has a name, it is marked next to that cluster. The C-terminal part of a representative cognate Phr sequence of each cluster is shown on the right. Underline marks the putative autoinducer peptide. The red letter marks the variable second residue of the putative autoinducer. (C) The YFP levels of a PspoIIG-YFP reporter integrated into strains over-expressing either RapK2-RR, RapK2-KR, or RapK2-KK. YFP was measured in the absence of peptide (gray) or following the addition of 10 μM of either ERPVGT (orange) or EKPVGT (red). See Methods and S5 Data for further details. Error bars mark standard errors. Asterisks mark cases in which peptide addition was significantly different from samples with no peptide. (D) Two possible evolutionary scenarios for the diversification of the RapK2 variants.