Figure 2. Ultrafast arsenic adaptation is due to rapid fixation of positively pleiotropic FPS1,ASK10 and ACR3 mutations.
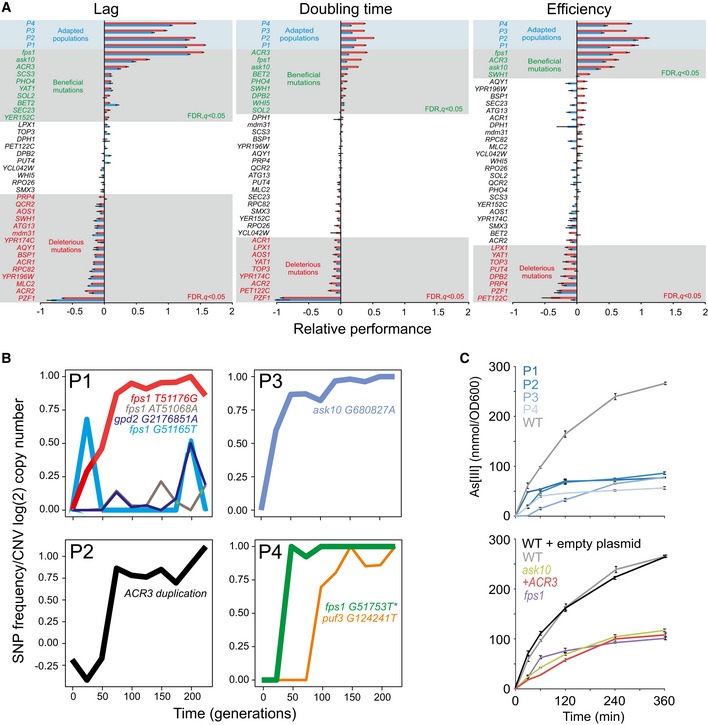
- Candidate driver mutations (gene duplications or SNP; see Materials and Methods) were reconstructed individually in a WT background and their growth in presence of As(III) evaluated. Mean (n = 2) fitness components in 3 and 5 mM As(III) (blue and red bars, respectively) relative to the founder (n = 20) are shown. Grey field = significant (FDR, q < 0.05) effects at 5 mM As(III). Blue field = As(III) adapted populations. Error bars represent SEM.
- Adapting P1–P4 populations were sampled at every 25th generation and deep sequenced. The frequency of confidently called mutations (total read depth of > 100, frequency of > 10% in ≥ 2 time points and snpEff effect = “Moderate” or “High”) predicted to affect protein function (SIFT < 0.05) is shown. Fps1 G51165T failed to pass the quality filter in the re‐sequencing but is shown for completeness. For the ACR3 containing duplicated region in P2, the grand copy number mean of all the segments within the duplicated region (chr. XVI 880799–944600) is shown. Colours indicate mutations. Bold line = causative mutations. *reconstructed FPS1 mutation.
- Arsenic accumulation inside cells. Top panel: Arsenic‐adapted populations (t = 250 generations). Bottom panel: As(III) causative mutations individually reconstructed in WT backgrounds. Error bars = SD (n = 2).
Source data are available online for this figure.
