Figure 4. RavJ modularity and inhibition by LegL1.
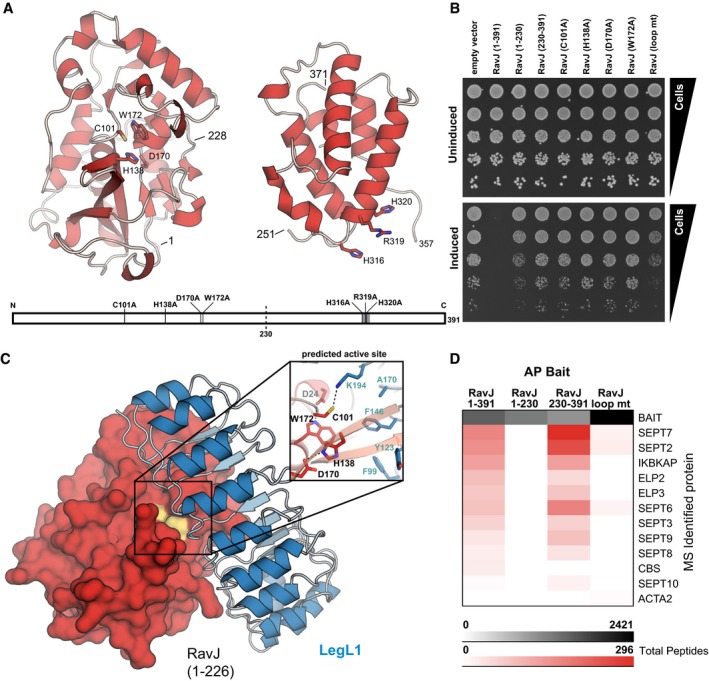
- Structures for amino acid residues 1–228 and 251–371 of RavJ were solved separately to a resolution of 1.3 and 1.9 Å, respectively. The N‐terminal structure contains a structurally conserved putative catalytic triad (C101/H138/D170). A conserved motif (highlighted in gray) is located in the C‐terminal domain with surface exposed residues H316, R319, and H320.
- While expression of full‐length RavJ showed a severe growth defect in a spot dilution assay, mutation of each of the putative catalytic residues or the adjacent W172 to alanine fully relieved toxicity as did substitutions in the conserved C‐terminal motif. Each mutant was tested for expression and stability (Appendix Fig S4C).
- LegL1 acts as a direct antagonist by blocking the active site of RavJ. The co‐crystal complex of LegL1 (blue) bound to RavJ (1–226, red) was solved to a resolution of 2.0 Å. LegL1 forms a canonical leucine‐rich repeat structure arching over the predicted active site of RavJ. The interface spans 1,240 Å2 and is reinforced by LegL1 K194 projecting into the RavJ active site where it forms a hydrogen bond with the predicted catalytic residue C101 (inset).
- Immobilized RavJ was incubated with U937 cell lysate and interacting proteins were identified by nLC‐MS/MS. Each column in the table represents the sum of the average total peptide counts for two replicates of affinity purification‐mass spectrometry. Septins and elongator complex proteins were identified only with full‐length RavJ and the (230–391) C‐terminal domain, suggesting that the C‐terminal domain is a substrate‐binding domain. Use of the otherwise full‐length loop mutant (H316A/R319A/H320A) as bait abrogated these interactions.
