Abstract
Background
The sortilin-related receptor 1 (SORL1) gene, regulating the trafficking and recycling of amyloid precursor protein, has been related to Alzheimer’s disease (AD) and mild cognitive impairment (MCI). The aim of the present study was to investigate the relationship between SORL1 polymorphisms, plasma concentrations of amyloid-beta (Aβ) isoforms, and AD and MCI susceptibility for a Han Chinese population in Taiwan.
Methods
Eight single-nucleotide polymorphisms (SNPs) in SORL1 and the apolipoprotein E gene (APOE) ε4 alleles were genotyped in 798 patients with AD, 157 patients with MCI, and 401 control subjects. Plasma concentrations of Aβ42, Aβ40, and neuropsychiatric tests for six different cognitive domains were examined.
Results
Among the eight tested SNPs, SORL1 rs1784933 was most significantly associated with AD and MCI in our population. The G allele of rs1784933 exerted a protective effect and was associated with a reduced risk of AD (odds ratio [OR] = 0.75, p = 0.004) and MCI (OR = 0.69, p = 0.013). The significance remained after we adjusted for age, sex, and APOE ε4 alleles. For the overall participants, the plasma concentrations of Aβ42 were nominally significant for subjects carrying the rs1784933 G allele having a lower level than those without the G allele (p = 0.046). There was a similar trend for the G allele carriers to have a lower plasma Aβ40 level than noncarriers, but this was not significant. The nonsynonymous SNP rs2298813 was also related to a lower disease risk when AD and MCI were combined as a group (OR = 0.76, p = 0.035). However, there was no association between SORL1 genotypes and any of the six cognitive tests.
Conclusions
Findings from our study provide support for the effect of SORL1 gene on the disease risks and pathognomonic surrogates of AD/MCI. The interaction between SORL1 polymorphisms and Aβ formation requires further study.
Keywords: SORL1 gene, Amyloid-beta, Plasma biomarkers, Polymorphisms, Alzheimer’s disease
Background
Alzheimer’s disease (AD) is a complex neurodegenerative disease caused by a combination of genetic and environmental influences. The heritability of AD was estimated to be 58–79% in a twins study [1]; yet, long lists of contributory genes have not been fully elucidated. Mutations in the amyloid precursor protein (APP), presenilin 1 (PSEN1), and presenilin 2 (PSEN2) genes are responsible for autosomal dominant inheritance of AD [2]. Genome-wide association studies (GWAS) identified more than 20 susceptible genes for late-onset Alzheimer’s disease (LOAD), including BIN1, CR1, CLU, CD33, PICALM, and ABCA7 [3–5]. Among them, the apolipoprotein E gene (APOE) remains the major genetic risk factor of LOAD by posing a 2.6- to 3.2-fold risk in individuals with one copy of the APOE ε4 allele and a 14.9-fold risk in those with two copies of the ε4 allele [6, 7].
Accumulation of amyloid-beta (Aβ) peptide, the neurotoxic proteolytic derivative of APP, is central to the pathogenesis of AD. The causative genes of familiar AD (APP, PSEN1, and PSEN2) and the strongest genetic contributor to LOAD (APOE) are all involved in the production, transport, and clearance of Aβ [2, 8]. The protein encoded by sortilin-related receptor 1 gene (SORL1) determines the intracellular fate of APP to be recycled or drifted to the endosome-lysosome pathway to generate Aβ [9, 10]. Aberrant expression of SORL1 has been implicated in AD pathogenesis because the SORL1 protein was found to be downregulated in the brain tissue of patients with sporadic AD [11]. Rogaeva et al. first illustrated that single-nucleotide polymorphisms (SNPs) located within two clusters of the SORL1 genome (SNPs 8–10 and SNPs 23–25) were related to LOAD susceptibility [12]. This association was later replicated in several ethnic groups, including white, Japanese, Korean, and Chinese populations [13–16]. Previous studies showed that SORL1 polymorphisms were related to decreased cerebrospinal fluid (CSF) concentrations of Aβ42 and Aβ40, as well as reduced CSF levels of SORL1 protein [17–19]. However, the relationship between SORL1 polymorphisms and plasma biomarkers of Aβ has never been investigated. Recent studies demonstrated that SORL1 polymorphisms predict atrophy of AD-specific brain structure (i.e., hippocampal and parahippocampal gyri) in nondemented elderly persons [20], supporting involvement of SORL1 in the neurodegeneration of cognition-related regions. Investigating the influence of SORL1 polymorphisms on these clinical and biological endophenotypes could strengthen their pathogenic role in AD.
The aim of the present study was to elucidate whether SORL1 polymorphisms confer a risk of LOAD and mild cognitive impairment (MCI) in the Han Chinese population in Taiwan, as well as deciphering its effects on different cognitive domains. The influence of SORL1 polymorphisms on different Aβ isoforms in blood was also examined to give biological evidence for SORL1’s effects.
Methods
Subjects
A total of 798 patients with LOAD, 157 patients with MCI, and 401 unrelated healthy control subjects were enrolled from Taipei Veterans General Hospital and Taichung Veterans General Hospital, Taiwan. All participants were of Han Chinese descent and resided in Taiwan. The diagnosis of probable AD was made according to the criteria of the National Institute of Neurological and Communicative Disorders and Stroke/Alzheimer’s Disease and Related Disorders Association [21], and the diagnosis of MCI was made according to the revised 2004 consensus criteria [22]. All participants received a comprehensive assessment, including history query, neurological examinations, laboratory tests, and neuroimaging as diagnostic surveys. The control subjects were volunteers without complaints of cognitive dysfunction recruited from outpatient clinics. The study was approved by the institutional review boards of each hospital. All participants provided informed consent according to our institutional guidelines and the recommendations of the Declaration of Helsinki.
Cognitive testing
For each participant, the global cognitive performance was assessed using the Mini Mental State Examination (MMSE) [23]. Tests specific to each cognitive domain were performed in patients with AD and patients with MCI, including (1) attention (forward and backward digit span from the Wechsler Memory Scale-IV) [24], (2) memory (12-item word recall test) [25], (3) language and executive function (verbal fluency category test) [26], (4) processing speed (Trail Making Test A) [27], and (5) naming task (Boston Naming Test) [28].
Genotyping
Genomic DNA was extracted from whole blood using the Gentra Puregene kit according to the manufacturer’s protocols (QIAGEN, Hilden, Germany). The ε2, ε3, and ε4 alleles of APOE were determined by two SNPs (rs429358 and rs7412) [29]. Eight SORL1 SNPs were selected on the basis of (1) rs2070045, rs1699102, rs3824968, rs2282649, and rs1010159 (aka SNP19, 22, 23, 24, and 25 in the original report by Rogaeva et al. [12]) being the top signals related to LOAD in white populations [12, 14]; (2) rs3737529 and rs1784933 being the most significant SNPs in Asian populations [15, 16]; and (3) the nonsynonymous SNP rs2298813 having been demonstrated to increase Aβ production in cellular models [30]. All genotyping reactions were carried out using the TaqMan genotyping assay (Applied Biosystems, Foster City, CA, USA). Polymerase chain reactions were performed using 96-well microplates with an ABI 7500 real-time polymerase chain reaction system (Applied Biosystems). Allele discrimination was achieved by detecting fluorescence using SDS software version 1.2.3 (Applied Biosystems).
Measurement of plasma Aβ concentrations
Plasma samples were available for 592 patients with LOAD, 119 patients with MCI, and 170 control subjects. Within 30 minutes of collection, plasma samples in ethylenediaminetetraacetic acid-containing tubes were centrifuged at 3000 rpm at 4 °C, and the supernatants were collected, divided into aliquots, and frozen at −80 °C. Plasma concentrations of Aβ40 and Aβ42 were measured using the INNO-BIA plasma Aβ forms immunoassay (Fujirebio, Gent, Belgium) as described previously [31]. In brief, the different Aβ isoforms were captured by a mix of beads selectively coated with three different monoclonal antibodies with affinity for Aβ42, Aβ40, and non-Aβ peptides. The immunoreactivity of Aβ42 and Aβ40 were quantified using the xMAP Technology on the Luminex analytical platform (Luminex, Austin, TX, USA).
Statistical analysis
Hardy-Weinberg equilibrium tests were conducted for each SNP. An additive model of inheritance was presumed to test the associations among SORL1 SNPs, LOAD, and MCI. A χ2 test was used to compare the genotype distributions between LOAD and control subjects, as well as between MCI and control subjects. Multivariate logistic regression without and with adjustment for age, sex, and APOE ε4 allele was further used to estimate the odds ratios (ORs) for the risk alleles.
To explore the influence of SOLR1 SNPs on AD endophenotypes, MMSE scores and serum Aβ concentrations were compared across different SORL1 genotypes using one-way analysis of variance (ANOVA). For patients with LOAD and patients with MCI, the influence of SORL1 genotypes on different cognitive domains was also evaluated using ANOVA. All statistical analyses were performed with PASW Statistics software (version 18.0; SPSS, Chicago, IL, USA) with a p value <0.05 set as statistically significant. Linkage disequilibrium (LD) blocks were generated by using Haploview version 5.0 software (Broad Institute, Cambridge, MA, USA) using the “solid spine of LD” method, in which a block was formed if the first and last markers were in strong LD with all intermediate markers. The frequency of each haplotype and comparison of the haplotype distributions between AD plus MCI in combination and the control group were performed using Haploview software version 5.0 [32]. To illustrate the LD conformation and haplotype frequency in white populations, the genotype data of the eight SORL1 SNPs from a CEU population (i.e., Utah residents with Northern and Western European ancestry) were obtained from the 1000 Genomes Project Browser (http://browser.1000genomes.org/index.html).
Results
Associations of SORL1 SNPs and AD/MCI risk
The demographic data of study participants are shown in Table 1. Eight SORL1 SNPs were genotyped, namely rs2298813 (SNP13), rs2070045 (SNP19), rs1699102 (SNP22), rs3824968 (SNP23), rs3737529, rs2282649 (SNP24), rs1010159 (SNP25), and rs1784933 (SNP26) (Table 2). The genotype distributions of all SNPs complied with Hardy-Weinberg equilibrium.
Table 1.
Demographic data
| Control subjects (n = 401) | MCI (n = 157) | AD (n = 798) | |
|---|---|---|---|
| Age, years | 75.4 ± 9.8 | 74.2 ± 8.3 | 79.1 ± 8.2** |
| Male sex | 257 (64.1%) | 82 (52.2%)* | 411 (51.5%)** |
| Education level, years | 11.1 ± 4.9 | 10.2 ± 4.7 | 9.7 ± 4.7** |
| MMSE score | 28.0 ± 2.1 | 26.0 ± 2.8** | 18.3 ± 5.9** |
| 12-item word recall test | – | 4.5 ± 2.9 | 1.4 ± 2.2† |
| Forward digit span | – | 10.0 ± 2.5 | 8.4 ± 3.0† |
| Backward digit span | – | 5.7 ± 2.4 | 3.9 ± 2.1† |
| Verbal fluency test | – | 10.2 ± 3.0 | 6.5 ± 3.2† |
| Boston Naming Test | – | 13.6 ± 1.3 | 11.4 ± 3.0† |
| Trail Making Test A, seconds | – | 92.3 ± 56.9 | 181.8 ± 145.2† |
| APOE genotypes | |||
| ε2ε2/ε2ε3/ε3ε3 | 339 (85.0%) | 123 (79.4%)* | 504 (63.3%)** |
| ε2ε4/ε3ε4 | 59 (14.8%) | 28 (18.1%) | 269 (33.8%) |
| ε4ε4 | 1 (0.3%) | 4 (2.6%) | 23 (2.9%) |
Abbreviations: MCI Mild cognitive impairment, AD Alzheimer’s disease, MMSE Mini Mental State Examination, APOE apolipoprotein E gene
Data are presented as count (percent) or mean (SD)
** p < 0.01 by χ2 test or Student’s t test when AD or MCI group was compared with control subjects
* p < 0.05 by χ2 test or Student’s t test when AD or MCI group was compared with control subjects
† p < 0.01 by Student’s t test when comparing patients with AD with patients with MCI
Table 2.
Genotype distribution of SORL1 single-nucleotide polymorphisms among patients with Alzheimer’s disease, patients with mild cognitive impairment, and control subjects
| MM/Mm/mm, n (%) | Multivariate regression analysis | ||||||
|---|---|---|---|---|---|---|---|
| SNP | Allele (M/m) | Control subjects | MCI | AD | MCI vs. control subjects | AD vs. control subjects | AD + MCI vs. control subjects |
| rs2298813 (SNP13) | G/A | 291/90/9 (74.6/23.1/2.3) | 134/18/2 (87.0/11.7/1.3) | 597/176/12 (76.1/22.4/1.5) | OR = 0.49, p = 0.003 | OR = 0.91, p = 0.455 | OR = 0.84, p = 0.154 |
| Adj OR = 0.49, p = 0.003 | Adj OR = 0.82, p = 0.156 | Adj OR = 0.76, p = 0.035 | |||||
| rs2070045 (SNP19) | G/T | 144/186/64 (36.5/47.2/16.2) | 54/77/25 (34.6/49.4/16.0) | 302/392/99 (38.1/49.4/12.5) | OR = 1.04, p = 0.794 | OR = 0.89, p = 0.204 | OR = 0.91, p = 0.307 |
| Adj OR = 1.02, p = 0.909 | Adj OR = 0.87, p = 0.149 | Adj OR = 0.90, p = 0.256 | |||||
| rs1699102 (SNP22) | C/T | 330/61/1 (84.2/15.6/0.3) | 129/28/0 (82.2/17.8/0.0) | 666/123/7 (83.7/15.5/0.9) | OR = 1.13, p = 0.620 | OR = 1.08, p = 0.637 | OR = 1.09, p = 0.596 |
| Adj OR = 1.13, p = 0.633 | Adj OR = 1.06, p = 0.711 | Adj OR = 1.07, p = 0.687 | |||||
| rs3824968 (SNP23) | A/T | 155/184/56 (39.2/46.6/14.2) | 61/76/19 (39.1/48.7/12.2) | 313/390/88 (39.6/49.3/11.1) | OR = 0.96, p = 0.772 | OR = 0.93, p = 0.409 | OR = 0.93, p = 0.431 |
| Adj OR = 0.95, p = 0.710 | Adj OR = 0.90, p = 0.300 | Adj OR = 0.92, p = 0.375 | |||||
| rs3737529 | C/T | 239/142/20 (59.6/35.4/5.0) | 103/50/4 (65.6/31.8/2.5) | 508/257/33 (63.7/32.2/4.1) | OR = 0.77, p = 0.120 | OR = 0.86, p = 0.164 | OR = 0.85, p = 0.107 |
| Adj OR = 0.78, p = 0.140 | Adj OR = 0.82, p = 0.073 | Adj OR = 0.82, p = 0.069 | |||||
| rs2282649 (SNP24) | T/C | 148/188/56 (37.8/48.0/14.3) | 59/79/18 (37.8/50.6/11.5) | 305/391/93 (38.7/49.6/11.8) | OR = 0.94, p = 0.659 | OR = 0.93, p = 0.409 | OR = 0.93, p = 0.408 |
| Adj OR = 0.92, p = 0.585 | Adj OR = 0.90, p = 0.297 | Adj OR = 0.92, p = 0.356 | |||||
| rs1010159 (SNP25) | C/T | 155/181/55 (39.6/46.3/14.1) | 62/77/18 (39.5/49.0/11.5) | 315/386/89 (39.9/48.9/11.3) | OR = 0.95, p = 0.702 | OR = 0.93, p = 0.462 | OR = 0.94, p = 0.463 |
| Adj OR = 0.93, p = 0.598 | Adj OR = 0.91, p = 0.322 | Adj OR = 0.92, p = 0.380 | |||||
| rs1784933 (SNP26) | A/G | 178/175/47 (44.5/43.8/11.8) | 86/61/10 (54.8/38.9/6.4) | 408/326/63 (51.2/40.9/7.9) | OR = 0.69, p = 0.012 | OR = 0.78, p = 0.008 | OR = 0.77, p = 0.003 |
| Adj OR = 0.69, p = 0.013 | Adj OR = 0.75, p = 0.004 | Adj OR = 0.74, p = 0.002 | |||||
Abbreviations: AD Alzheimer’s disease, MCI Mild cognitive impairment, SNP Single-nucleotide polymorphism, SORL1 Sortilin-related receptor 1 gene, M Major allele, m Minor allele, OR Odds ratio
Model of inheritance was an additive model. Adjusted ORs and adjusted p values were obtained from logistic regression with adjustment of age, sex, and APOE ε4 allele
SORL1 rs1784933 was most significantly associated with LOAD and MCI in our population (Table 2). The minor allele G of rs1784933 appeared to exert a protective effect, with significantly lower frequencies of GG genotype in patients with LOAD (7.9%) and in patients with MCI (6.4%) in comparison with control subjects (11.8%) (Table 2). In regression analysis, the G allele was associated with a reduced risk of MCI and LOAD. After adjustment for age, sex, and APOE ε4 allele, the G allele remained a significant predictor of MCI (OR = 0.69, p = 0.013) and LOAD (OR = 0.75, p = 0.004). Combing patients with AD and patients with MCI revealed a stronger association between SORL1 rs1784933 and AD spectrum disorder (OR = 0.74, p = 0.002).
In addition to rs1784933, the nonsynonymous SNP rs2298813 was also significantly associated with MCI (Table 2). The minor allele A of rs2298813 carried a reduced risk of MCI (OR = 0.49, p = 0.003) and remained significant after adjustment for age, sex, and APOE ε4 allele. The A allele of rs2298813 showed a similar trend, with a protective effect on LOAD, but this result was insignificant. When patients with AD and patients with MCI were combined, the minor allele A of rs2298813 was significantly associated with a reduced risk after adjustment for other covariates (OR = 0.76, p = 0.035).
Haplotype analysis
LD mapping of the eight genotyped SORL1 SNPs showed that there were two LD blocks in the Han Chinese population in Taiwan (Fig. 1, left panel). One LD block was composed of rs2070045 and rs1699102 (SNPs 19–22), and the other one included four SNPs (rs3824968, rs3737529, rs2282649, and rs1010159; SNPs 23–rs3737529–25). Haplotype analysis was performed in the two LD blocks separately, but it failed to yield any significant result (Fig. 1). When comparing the LD maps between the Han Chinese population in the present study and the CEU population from the 1000 Genomes Project, the LD conformation and the haplotype frequency substantially differed between the two ethnic groups. For example, “ACTC” was the most common haplotype of the second LD block (SNPs 23–rs3737529–SNP25) in the Han Chinese population, but “TCCT” was the most common one in the CEU population (Fig. 1).
Fig. 1.
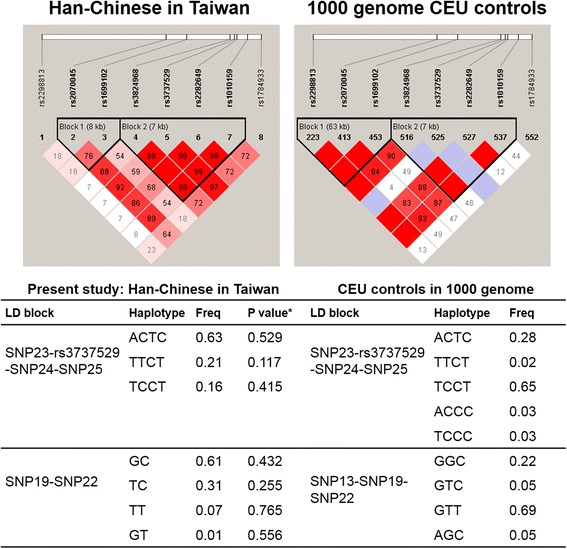
Linkage disequilibrium (LD) map and haplotype analysis. * p value for comparing haplotype distribution between patients with AD + patients with MCI and control subjects using Haploview software version 5.0. The frequency (Freq) of each haplotype and the conformation of LD blocks differed substantially between Han Chinese in Taiwan and a white population (CEU; Utah residents with Northern and Western European ancestry) from the 1000 Genomes Project. SNP Single-nucleotide polymorphism
Associations of SORL1 SNPs and plasma Aβ concentrations
We further explored the relationship between rs1784933 genotypes and plasma Aβ concentrations (Table 3). The average plasma concentrations of Aβ40 and Aβ42, as well as the ratios of plasma Aβ42/Aβ40 concentration, were similar among control subjects, patients with MCI, and patients with LOAD. Overall, the plasma concentrations of Aβ40 and Aβ42 were significantly lower in subjects carrying the rs1784933 G allele than among those without that G allele. After adjusting for age, sex, and APOE ε4 allele, the association of the G allele of SORL1 rs1784933 with a lower plasma concentration of Aβ42 remained nominally significant (p = 0.046); however, the results were only borderline significant for a lower plasma concentration of Aβ40 (p = 0.071). In addition, the ratio of plasma Aβ42/Aβ40 concentration did not differ between rs1784933 G allele carriers and noncarriers. Similar analysis done for patients with LOAD alone revealed lower plasma Aβ42 concentrations in G allele carriers than in noncarriers. However, the plasma concentration of Aβ40 and the ratio of Aβ42/Aβ40 concentration in patients with LOAD did not show any difference between G allele carriers and noncarriers.
Table 3.
Associations between SORL1 rs1784933 and plasma amyloid-beta concentration
| Multivariate regression model | Aβ42 | Aβ40 | Aβ42/Aβ40 ratio | |
|---|---|---|---|---|
| AD (n = 592) | 23.8 ± 15.1 | 173.1 ± 79.3 | 0.15 ± 0.25 | |
| MCI (n = 119) | 23.6 ± 12.5 | 178.7 ± 54.6 | 0.14 ± 0.07 | |
| Control subjects (n = 170) | 23.7 ± 12.6 | 171.6 ± 64.3 | 0.15 ± 0.08 | |
| AD vs. control subjects | p = 0.899 | p = 0.807 | p = 0.904 | |
| MCI vs. control subjects | p = 0.969 | p = 0.318 | p = 0.189 | |
| SORL1 rs1784933 in overall participants (n = 873) | ||||
| G allele carriers (AG + GG genotypes) | 22.68 ± 13.82 | 168.51 ± 65.93 | 0.14 ± 0.08 | |
| G allele noncarriers (AA genotype) | 24.82 ± 14.67 | 178.33 ± 79.81 | 0.16 ± 0.27 | |
| G allele carriers vs. noncarriers | ||||
| Raw p value | p = 0.026 | p = 0.048 | p = 0.214 | |
| Adjusted for age and sex | Adj p = 0.029 | Adj p = 0.054 | Adj p = 0.217 | |
| Adjusted for age, sex, and APOE ε4 allele | Adj p = 0.046 | Adj p = 0.071 | Adj p = 0.248 | |
| SORL1 rs1784933 in patients with AD (n = 584) | ||||
| G allele carriers (AG + GG genotypes) | 22.62 ± 14.48 | 168.86 ± 70.79 | 0.14 ± 0.08 | |
| G allele noncarriers (AA genotype) | 25.00 ± 15.60 | 177.14 ± 86.24 | 0.16 ± 0.33 | |
| G allele carriers vs. noncarriers | ||||
| Raw p value | p = 0.056 | p = 0.205 | p = 0.203 | |
| Adjusted for age and sex | Adj p = 0.056 | Adj p = 0.204 | Adj p = 0.203 | |
| Adjusted for age, sex, and APOE ε4 allele | Adj p = 0.058 | Adj p = 0.211 | Adj p = 0.206 | |
Abbreviations: AD Alzheimer’s disease, APOE Apolipoprotein E gene, MCI Mild cognitive impairment, SORL1 Sortilin-related receptor 1 gene, Aβ Amyloid-beta, Adj p p value in the multivariate regression with adjustment of covariates
Associations of SORL1 SNPs and cognitive tests
To further test the influence of rs1784933 genotypes on cognitive function, the average MMSE scores among the three genotypes were compared in patients with LOAD, patients with MCI, and control subjects separately. There was no significant difference in the MMSE scores among the AA, AG, and GG genotypes of rs1784933 (Table 4). For patients with MCI and patients with LOAD, there was no association between rs1784933 genotypes and any of the six cognitive test results (Table 4). Neither the MMSE scores nor any of the six cognitive domain tests showed differences across rs2298813 genotypes (data not shown).
Table 4.
Associations between SORL1 rs1784933 and cognitive tests
| Subjects | Cognitive tests | SORL1 rs1784933 | p Value (ANOVA) | ||
|---|---|---|---|---|---|
| AA | AG | GG | |||
| Control subjects | MMSE score | 28.0 ± 2.1 | 28.1 ± 1.9 | 27.6 ± 2.1 | 0.490 |
| MCI | MMSE score | 25.9 ± 2.9 | 26.0 ± 2.8 | 26.7 ± 2.4 | 0.468 |
| 12-item word recall test | 4.5 ± 3.0 | 4.5 ± 2.7 | 5.0 ± 2.6 | 0.877 | |
| Forward digit span | 10.0 ± 2.8 | 10.2 ± 2.1 | 9.1 ± 2.9 | 0.507 | |
| Backward digit span | 5.5 ± 2.5 | 6.1 ± 2.1 | 6.3 ± 3.2 | 0.271 | |
| Verbal fluency test | 10.1 ± 2.8 | 10.5 ± 3.4 | 9.5 ± 2.3 | 0.576 | |
| Boston Naming Test | 13.5 ± 1.2 | 13.8 ± 1.3 | 13.4 ± 1.4 | 0.290 | |
| Trail Making Test A, seconds | 99.7 ± 67.2 | 84.1 ± 38.9 | 74.4 ± 39.0 | 0.177 | |
| AD | MMSE score | 18.1 ± 6.0 | 18.5 ± 5.7 | 18.5 ± 6.0 | 0.648 |
| 12-item word recall test | 1.3 ± 2.1 | 1.5 ± 2.2 | 1.5 ± 2.5 | 0.494 | |
| Forward digit span | 8.3 ± 3.1 | 8.6 ± 3.1 | 8.1 ± 2.2 | 0.224 | |
| Backward digit span | 3.8 ± 2.1 | 3.9 ± 2.2 | 3.5 ± 1.8 | 0.461 | |
| Verbal fluency test | 6.4 ± 3.3 | 6.5 ± 3.1 | 6.9 ± 2.8 | 0.537 | |
| Boston Naming Test | 11.2 ± 3.1 | 11.5 ± 2.9 | 11.9 ± 2.3 | 0.229 | |
| Trail Making Test A, seconds | 186.8 ± 160.5 | 175.6 ± 130.4 | 182.9 ± 115.2 | 0.630 | |
Abbreviations: AD Alzheimer’s disease, MCI Mild cognitive impairment, SORL1 Sortilin-related receptor 1 gene, MMSE Mini Mental State Examination, ANOVA One-way analysis of variance
Discussion
The present study confirmed SORL1 as a susceptible gene for LOAD and MCI in the Han Chinese population in Taiwan. The SNP rs1784933 located in the 3′ region of the SORL1 genome and the nonsynonymous SNP rs2298813 were most significantly associated with AD and MCI. A lower plasma level of Aβ42 was found in individuals carrying the minor allele G of rs1784933 in comparison with those without the G allele. A similar trend of reduced plasma levels of Aβ40 was also observed in the G allele carriers, but this finding was not significant. Neither MMSE scores nor any test of the six cognitive domains differed among SORL1 genotypes.
In the Taiwanese population, SNP rs1784933 (SNP26) is most significantly associated with AD/MCI susceptibility, and its minor allele G exerts a protective effect against disease. Consistent with our findings, in a study of persons of Han Chinese descent in mainland China, researchers found that the G allele of rs1784933, but not the other two tested SORL1 SNPs was related to a reduced risk of AD [16]. Although the associations between SORL1 polymorphisms and AD have been replicated in several studies [14, 15], the regions tagged by most significant SNPs vary across different ethnic groups. For whites, Caribbean Hispanics, and Israeli Arabs, SNPs located in the 5′ end of the SORL1 genome (i.e., SNPs 8–10) are most strongly associated with AD [12, 14]. However, SNPs near the 3′ region of the SORL1 genome (i.e., SNP19 and SNPs 22–25) are more significantly related to AD in the Chinese, Japanese, and African American populations [13, 33, 34]. The consistent findings between our study and other Asian groups imply a pathogenic role of the 3′ region of SORL1 in AD, especially for Asian populations. In addition, the different haplotype frequency and LD conformation between Han Chinese and CEU populations (Fig. 1) further explain why the most significant SNPs vary across populations.
It is worthwhile to note that the nonsynonymous SNP rs2298813 (A528T), causing an amino acid substitution from alanine to threonine at the 528th residue of SORL1 protein, was significantly associated with MCI in our population. A similar but insignificant effect of rs2298813 on LOAD was also observed. Interestingly, rs2298813 was rarely found significant in previous GWAS of LOAD, but this coding variant segregates with disease status in familial AD [30]. The results of an in vitro study suggest that this coding variant has a direct and deleterious impact on AD pathogenesis because HEK293 cells expressing A528T mutant SORL1 could not physiologically interact with APP, which subsequently increased the secretion of Aβ42, soluble APPα, and APPβ [30].
The SORL1 protein regulates APP trafficking and processing, which subsequently influences the formation of Aβ [9]. Researchers in several studies explored the relationship between SORL1 polymorphisms and CSF levels of Aβ42 and Aβ40, but their work led conflicting results [17–19, 35, 36]. Concordant with our findings that subjects carrying the minor allele of rs1784933 have reduced plasma levels of Aβ42, investigators in several studies found that SNPs located at the 3′ region of SORL1 were associated with lower concentrations of Aβ42 in CSF [17–19]. A trend for reduced CSF concentrations of Aβ40 was also observed in these studies, but without statistical significance. Because SORL1 regulates the APP processing pathway upstream from the catalyzation of β- and γ-secretases, insufficient SORL1 activity would not change the ratio of Aβ42/Aβ40 concentrations.
To our knowledge, the present study is the first investigation of the influence of SORL1 polymorphisms on plasma concentrations of Aβ42 and Aβ40. Although plasma Aβ concentrations might be confounded by age, disease duration, and other factors [37, 38], they are more easily accessible than CSF Aβ levels as a surrogate marker of AD pathology. Notably, the association between plasma Aβ levels and SORL1 rs1784933 derived mainly from patients with AD rather than from patients with MCI (Table 3). There might be two reasons for such a discrepancy. First, only 30–60% of patients with MCI have a neurodegenerative and progressive course, with the remainder having nondegenerative (or reversible) causes [39, 40]. The MCI group consists of heterogeneous entities, including AD and other pathogenesis, which might account for the insignificant correlation between SORL1 polymorphisms and plasma Aβ concentrations. Second, the smaller sample size of the patients with MCI with available plasma Aβ levels may have limited our power to detect a significant correlation. We did not measure the plasma concentrations of SORL1 protein, because it is undetectable in the circulation, according to a previous study [41].
The relationship between SORL1 variants and cognitive function has been investigated. Reynolds et al. found that markers at the 5′ region of SORL1 tended to be associated with verbal function decline and that SNPs near the 3′ end were more related with episodic memory impairment [42]. However, in a large cohort with a sample size up to 9624 participants, researchers did not find any correlation between SORL1 variants and different domains of cognitive function [43]. The present study also does not demonstrate any association between SORL1 SNPs and the six cognitive domains.
Conclusions
SORL1 is genetically related to MCI and LOAD in the Han Chinese population in Taiwan. A reduced plasma concentration of Aβ42 was found in individuals carrying the minor allele of the most significant SNP, rs1784933, implying a biological role of SORL1 genetic markers on the Aβ cascade.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from Academia Sinica of Taiwan (Taiwan Biobank, Stroke Biosignature Project study of Alzheimer disease); the Ministry of Science and Technology of Taiwan (104-2314-B-075-005-MY2, 104-2745-B-075-001-); Taipei Veterans General Hospital (V105C-110, V105E9-001-MY2-1); Ministry of Science and Technology (Republic of China) support for the Center for Dynamical Biomarkers and Translational Medicine, National Central University, Taiwan (MOST 103-2911-I-008-001); the Brain Research Center, National Yang-Ming University; and the Ministry of Education Aim for the Top University Plan.
Availability of supporting data
Not applicable.
Authors’ contributions
CTC participated in data analysis and drafting of the manuscript. YCL participated in genotyping, haplotype analysis, and revision of the manuscript. WJL participated in study design, collecting the data, and performing plasma marker measurements. SJW contributed to study design, acquisition of data, and obtaining funding. JLF conceived of the study and contributed to study coordination, acquisition of data, obtaining funding, and revision of the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
All the authors gave their approval for publication.
Ethics approval and consent to participate
This research was approved by the local ethics committees at Taipei Veterans General Hospital (IRB number 2012-05-033B). All subjects or, where appropriate, their nearest relatives provided written informed consent to participate.
Abbreviations
- AD
Alzheimer’s disease
- ANOVA
Analysis of variance
- APOE
Apolipoprotein E gene
- APP
Amyloid precursor protein gene
- Aβ
Amyloid-beta
- CEU
Utah residents with Northern and Western European ancestry
- CSF
Cerebrospinal fluid
- GWAS
Genome-wide association studies
- LD
Linkage disequilibrium
- LOAD
Late-onset Alzheimer’s disease
- MCI
Mild cognitive impairment
- MMSE
Mini Mental State Examination
- PSEN1
Presenilin 1 gene
- PSEN2
Presenilin 2 gene
- SNP
Single-nucleotide polymorphism
- SORL1
Sortilin-related receptor 1 gene
Contributor Information
Cheng-Ta Chou, Email: ctchou2@vghtc.gov.tw.
Yi-Chu Liao, Email: yichu.liao@gmail.com.
Wei-Ju Lee, Email: weijlee@vghtc.gov.tw.
Shuu-Jiun Wang, Phone: 886-2-28762522, Email: sjwang@vghtpe.gov.tw.
Jong-Ling Fuh, Phone: 886-2-28762522, Email: stellafuh@gmail.com, Email: jlfuh@vghtpe.gov.tw.
References
- 1.Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63:168–74. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 2.Cacace R, Sleegers K, Van Broeckhoven C. Molecular genetics of early-onset Alzheimer’s disease revisited. Alzheimers Dement. 2016;12:733–48. doi: 10.1016/j.jalz.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 3.Bertram L, Lill CM, Tanzi RE. The genetics of Alzheimer disease: back to the future. Neuron. 2010;68:270–81. doi: 10.1016/j.neuron.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 4.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan L, Yu JT, Zhang W, Wu ZC, Zhang Q, Liu QY, et al. Association of GWAS-linked loci with late-onset Alzheimer’s disease in a northern Han Chinese population. Alzheimers Dement. 2013;9:546–53. doi: 10.1016/j.jalz.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–3. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 7.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease: a meta-analysis. JAMA. 1997;278:1349–56. doi: 10.1001/jama.1997.03550160069041. [DOI] [PubMed] [Google Scholar]
- 8.Selkoe DJ. Alzheimer’s disease: genes, proteins, and therapy. Physiol Rev. 2001;81:741–66. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 9.Andersen OM, Reiche J, Schmidt V, Gotthardt M, Spoelgen R, Behlke J, et al. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci U S A. 2005;102:13461–6. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dodson SE, Andersen OM, Karmali V, Fritz JJ, Cheng D, Peng J, et al. Loss of LR11/SORLA enhances early pathology in a mouse model of amyloidosis: evidence for a proximal role in Alzheimer’s disease. J Neurosci. 2008;28:12877–86. doi: 10.1523/JNEUROSCI.4582-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dodson SE, Gearing M, Lippa CF, Montine TJ, Levey AI, Lah JJ. LR11/SorLA expression is reduced in sporadic Alzheimer disease but not in familial Alzheimer disease. J Neuropathol Exp Neurol. 2006;65:866–72. doi: 10.1097/01.jnen.0000228205.19915.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–77. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura R, Yamamoto M, Morihara T, Akatsu H, Kudo T, Kamino K, et al. SORL1 is genetically associated with Alzheimer disease in a Japanese population. Neurosci Lett. 2009;461:177–80. doi: 10.1016/j.neulet.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 14.Reitz C, Cheng R, Rogaeva E, Lee JH, Tokuhiro S, Zou F, et al. Meta-analysis of the association between variants in SORL1 and Alzheimer disease. Arch Neurol. 2011;68:99–106. doi: 10.1001/archneurol.2010.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyashita A, Koike A, Jun G, Wang LS, Takahashi S, Matsubara E, et al. SORL1 is genetically associated with late-onset Alzheimer’s disease in Japanese, Koreans and Caucasians. PLoS One. 2013;8:e58618. doi: 10.1371/journal.pone.0058618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng X, Hou D, Deng Y, Li W, Tian M, Yu Z. SORL1 gene polymorphism association with late-onset Alzheimer’s disease. Neurosci Lett. 2015;584:382–9. doi: 10.1016/j.neulet.2014.10.055. [DOI] [PubMed] [Google Scholar]
- 17.Kolsch H, Jessen F, Wiltfang J, Lewczuk P, Dichgans M, Kornhuber J, et al. Influence of SORL1 gene variants: association with CSF amyloid-beta products in probable Alzheimer’s disease. Neurosci Lett. 2008;440:68–71. doi: 10.1016/j.neulet.2008.05.049. [DOI] [PubMed] [Google Scholar]
- 18.Alexopoulos P, Guo LH, Kratzer M, Westerteicher C, Kurz A, Perneczky R. Impact of SORL1 single nucleotide polymorphisms on Alzheimer’s disease cerebrospinal fluid markers. Dement Geriatr Cogn Disord. 2011;32:164–70. doi: 10.1159/000332017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo LH, Westerteicher C, Wang XH, Kratzer M, Tsolakidou A, Jiang M, et al. SORL1 genetic variants and cerebrospinal fluid biomarkers of Alzheimer’s disease. Eur Arch Psychiatry Clin Neurosci. 2012;262:529–34. doi: 10.1007/s00406-012-0295-x. [DOI] [PubMed] [Google Scholar]
- 20.Yin RH, Li J, Tan L, Wang HF, Tan MS, Yu WJ, et al. Impact of SORL1 genetic variations on MRI markers in non-demented elders. Oncotarget. 2016;7:31689–98. doi: 10.18632/oncotarget.9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34:939–44. doi: 10.1212/WNL.34.7.939. [DOI] [PubMed] [Google Scholar]
- 22.Winblad B, Palmer K, Kivipelto M, Jelic V, Fratiglioni L, Wahlund LO, et al. Mild cognitive impairment – beyond controversies, towards a consensus: report of the International Working Group on Mild Cognitive Impairment. J Intern Med. 2004;256:240–6. doi: 10.1111/j.1365-2796.2004.01380.x. [DOI] [PubMed] [Google Scholar]
- 23.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”: a practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–98. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 24.Wechsler D. WMS-IV: Wechsler Memory Scale. 4. San Antonio, TX: The Psychological Corporation; 2009. [Google Scholar]
- 25.Vanderploeg RD, Schinka JA, Jones T, Small BJ, Graves AB, Mortimer JA. Elderly norms for the Hopkins Verbal Learning Test-Revised. Clin Neuropsychol. 2000;14:318–24. doi: 10.1076/1385-4046(200008)14:3;1-P;FT318. [DOI] [PubMed] [Google Scholar]
- 26.Harrison JE, Buxton P, Husain M, Wise R. Short test of semantic and phonological fluency: normal performance, validity and test-retest reliability. Br J Clin Psychol. 2000;39:181–91. doi: 10.1348/014466500163202. [DOI] [PubMed] [Google Scholar]
- 27.Lu L, Bigler ED. Normative data on Trail Making Test for neurologically normal, Chinese-speaking adults. Appl Neuropsychol. 2002;9:219–25. doi: 10.1207/S15324826AN0904_4. [DOI] [PubMed] [Google Scholar]
- 28.Mack WJ, Freed DM, Williams BW, Henderson VW. Boston Naming Test: shortened versions for use in Alzheimer’s disease. J Gerontol. 1992;47:P154–158. doi: 10.1093/geronj/47.3.P154. [DOI] [PubMed] [Google Scholar]
- 29.Chen CS, Ouyang P, Yeh YC, Lai CL, Liu CK, Yen CF, et al. Apolipoprotein E polymorphism and behavioral and psychological symptoms of dementia in patients with Alzheimer disease. Alzheimer Dis Assoc Disord. 2012;26:135–9. doi: 10.1097/WAD.0b013e31821f5787. [DOI] [PubMed] [Google Scholar]
- 30.Vardarajan BN, Zhang Y, Lee JH, Cheng R, Bohm C, Ghani M, et al. Coding mutations in SORL1 and Alzheimer disease. Ann Neurol. 2015;77:215–27. doi: 10.1002/ana.24305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lachno DR, Vanderstichele H, De Groote G, Kostanjevecki V, De Meyer G, Siemers ER, et al. The influence of matrix type, diurnal rhythm and sample collection and processing on the measurement of plasma beta-amyloid isoforms using the INNO-BIA plasma Aβ forms multiplex assay. J Nutr Health Aging. 2009;13:220–5. doi: 10.1007/s12603-009-0062-5. [DOI] [PubMed] [Google Scholar]
- 32.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 33.Lee JH, Cheng R, Schupf N, Manly J, Lantigua R, Stern Y, et al. The association between genetic variants in SORL1 and Alzheimer disease in an urban, multiethnic, community-based cohort. Arch Neurol. 2007;64:501–6. doi: 10.1001/archneur.64.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan EK, Lee J, Chen CP, Teo YY, Zhao Y, Lee WL. SORL1 haplotypes modulate risk of Alzheimer’s disease in Chinese. Neurobiol Aging. 2009;30:1048–51. doi: 10.1016/j.neurobiolaging.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 35.Kauwe JS, Cruchaga C, Bertelsen S, Mayo K, Latu W, Nowotny P, et al. Validating predicted biological effects of Alzheimer’s disease associated SNPs using CSF biomarker levels. J Alzheimers Dis. 2010;21:833–42. doi: 10.3233/JAD-2010-091711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elias-Sonnenschein LS, Helisalmi S, Natunen T, Hall A, Paajanen T, Herukka SK, et al. Genetic loci associated with Alzheimer’s disease and cerebrospinal fluid biomarkers in a Finnish case-control cohort. PLoS One. 2013;8:e59676. doi: 10.1371/journal.pone.0059676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayeux R, Honig LS, Tang MX, Manly J, Stern Y, Schupf N, et al. Plasma Aβ40 and Aβ42 and Alzheimer’s disease: relation to age, mortality, and risk. Neurology. 2003;61:1185–90. doi: 10.1212/01.WNL.0000091890.32140.8F. [DOI] [PubMed] [Google Scholar]
- 38.Sundelof J, Giedraitis V, Irizarry MC, Sundstrom J, Ingelsson E, Ronnemaa E, et al. Plasma beta amyloid and the risk of Alzheimer disease and dementia in elderly men: a prospective, population-based cohort study. Arch Neurol. 2008;65:256–63. doi: 10.1001/archneurol.2007.57. [DOI] [PubMed] [Google Scholar]
- 39.Burns JM, Bennett DA. Parsing the heterogeneity of mild cognitive impairment: lumpers and splitters. Neurology. 2015;85:1646–7. doi: 10.1212/WNL.0000000000002110. [DOI] [PubMed] [Google Scholar]
- 40.Langa KM, Levine DA. The diagnosis and management of mild cognitive impairment: a clinical review. JAMA. 2014;312:2551–61. doi: 10.1001/jama.2014.13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma QL, Galasko DR, Ringman JM, Vinters HV, Edland SD, Pomakian J, et al. Reduction of SorLA/LR11, a sorting protein limiting beta-amyloid production, in Alzheimer disease cerebrospinal fluid. Arch Neurol. 2009;66:448–57. doi: 10.1001/archneurol.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reynolds CA, Zavala C, Gatz M, Vie L, Johansson B, Malmberg B, et al. Sortilin receptor 1 predicts longitudinal cognitive change. Neurobiol Aging. 2013;1710(34):e1711–1718. doi: 10.1016/j.neurobiolaging.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu F, Ikram MA, Janssens AC, Schuur M, de Koning I, Isaacs A, et al. A study of the SORL1 gene in Alzheimer’s disease and cognitive function. J Alzheimers Dis. 2009;18:51–64. doi: 10.3233/JAD-2009-1137. [DOI] [PubMed] [Google Scholar]


