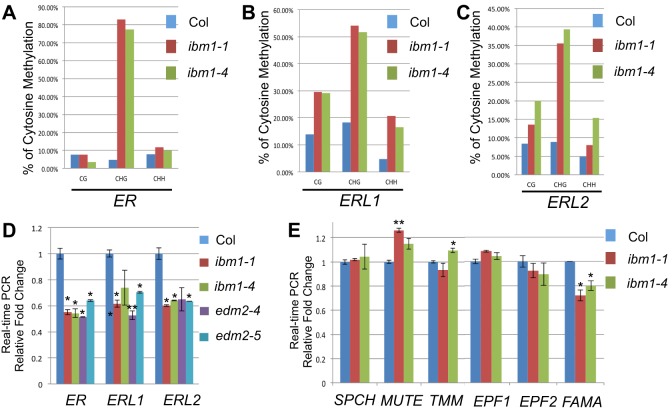
Fig. 5.
The ibm1 mutations affect the DNA methylation levels of ER genes. (A-C) Individual locus bisulfite sequencing analyses of CG, CHG, CHH methylation in ER (A), ERL1 (B) and ERL2 (C) in Col, ibm1-1 and ibm1-4. At least ten clones were selected from each sample. Striking methylation elevation in CHGs was found in all three ER genes when IBM1 is defective. (D,E) Transcript level of stomata-related genes in Col and in ibm1 and edm2 mutants. The expression of ER genes was suppressed in ibm1 and edm2 mutants. Values are mean±s.d. Data were collected from three biological replicates and the expression fold changes were normalized to the transcript level in Col. *P<0.05, **P<0.01, compared with the wild type (Col) by Student's t-test with Bonferroni correction; n=3 replicates.