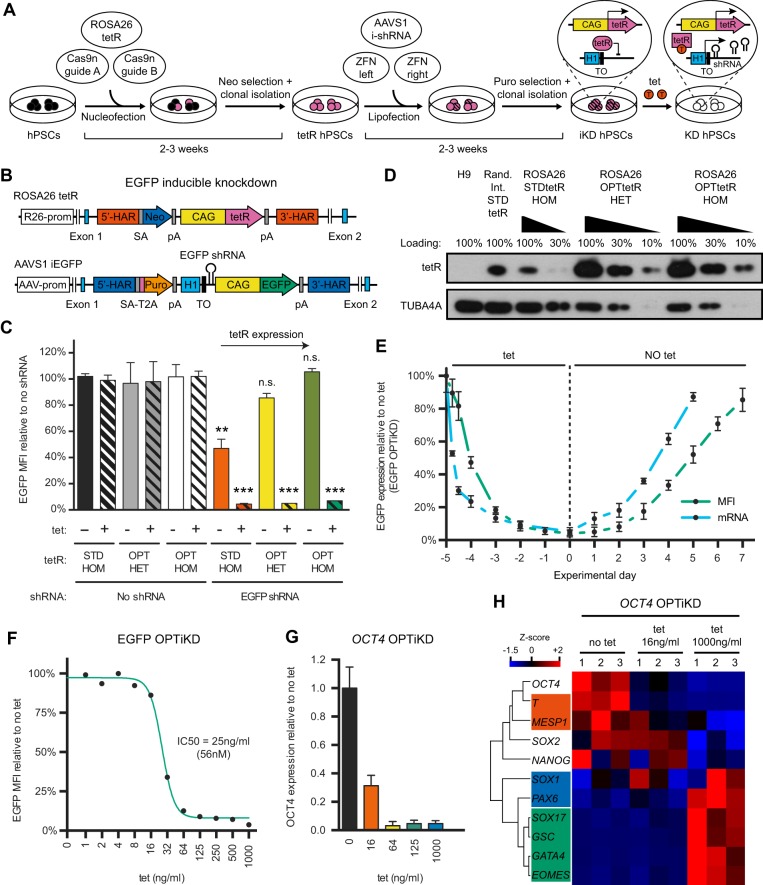
Fig. 2.
Development of an optimized inducible knockdown system (OPTiKD) based on dual GSH targeting of hPSCs. (A) Experimental approach for the generation of inducible knockdown (iKD) hPSCs. H1, H1 promoter; TO, tet operon; tetR, tetracycline-controlled repressor; ZFN, zinc-finger nuclease. (B) Transgenic alleles generated to obtain hESCs expressing an EGFP reporter transgene that could be silenced using an inducible EGFP shRNA. (C) EGFP expression in the absence or presence of tetracycline for 5 days in hESCs targeted with the indicated combinations of inducible EGFP shRNA and tetR [wild-type standard tetR (STDtetR) or codon-optimized tetR (OPTtetR)]. Double-targeted hESCs that did not carry the EGFP shRNA were used as negative controls. Results are from two or three individual lines per condition (see Table S1). n.s., P>0.05 (non-significant), **P<0.01, ***P<0.001 versus the same tetR line no tet and no shRNA (ANOVA with post-hoc Holm-Sidak comparisons). (D) Representative western blot for tetR in ROSA26-targeted hESCs expressing STDtetR or OPTtetR. HET, heterozygous targeting; HOM, homozygous targeting. hESCs with STDtetR random integration (Rand. Int.) are shown as a positive reference, while wild-type H9 hESCs are negative controls. Various amounts of protein were loaded to facilitate semi-quantitative comparison. TUBA4A (α-tubulin) provided a loading control. (E) EGFP knockdown and rescue kinetics in EGFP OPTiKD hESCs measured by flow cytometry (MFI) and qPCR (mRNA). Results are from two independent cultures per time point. (F) Tetracycline dose-response curve for EGFP knockdown in EGFP OPTiKD hESCs. The half-maximal inhibitory concentration (IC50) is reported. Results are from two independent cultures per dose, and the mean is shown. (G,H) qPCR analysis of OCT4 OPTiKD hESCs in the absence of tetracycline, or following treatment with different doses of tetracycline for 5 days. (H) Genes are clustered by complete Euclidean distance, and genes specific for pluripotency or for the primary germ layers are in color-coded boxes: no color, hPSCs; red, mesoderm; green, endoderm; blue, neuroectoderm. Z-scores indicate differential expression measured in the number of standard deviations from the average level. Results are from three independent cultures.