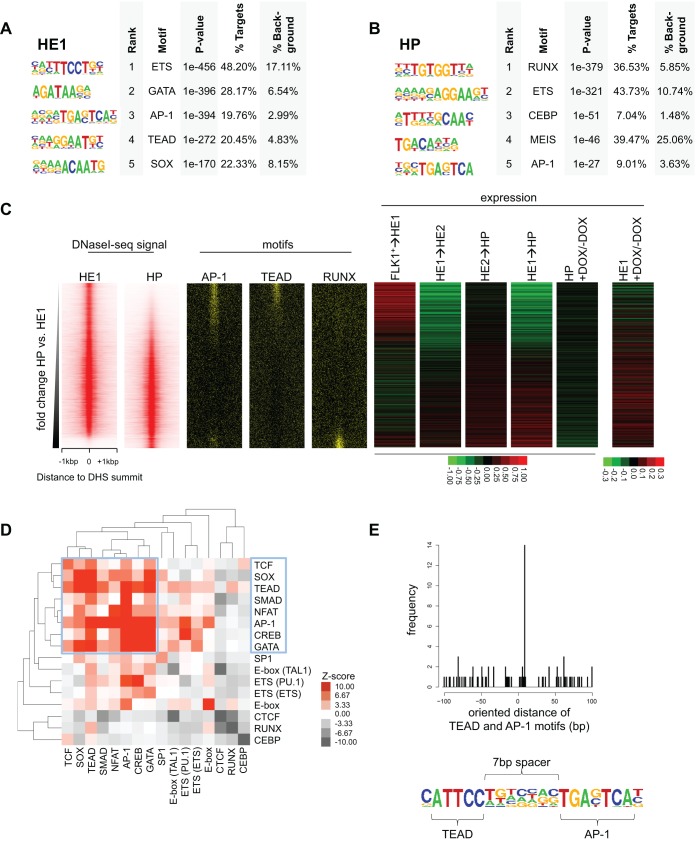
Fig. 7.
AP-1 and TEAD footprints occur together in HE1 DHS peaks and are strongly reduced in HP hypersensitive sites. (A) Transcription factor-binding motifs enriched in HE1-specific DHSs employing HOMER de novo motif discovery. (B) As in A, examining HP-specific DHSs. (C) Heatmaps depicting HE1 and HP DNaseI-seq signal (left), motif (middle) and gene expression fold change (right) sorted based on increasing HP/HE1 fold change. (D) Co-occurrence clustering of factor binding motifs specifically occupied (footprinted) in HE1 demonstrating the co-occurrence of motifs for signalling-responsive transcription factors. Z-scores represent enrichment over 1000 equally sized, random sub-samplings in HP DHSs. (E) AP-1 and TEAD motifs are arranged in a specific conformation. Distribution of AP-1 motif start distances to TEAD motif starts, aligned by TEAD motif orientation (top). Bottom: composite TEAD-AP1 motif with 7 bp spacer.