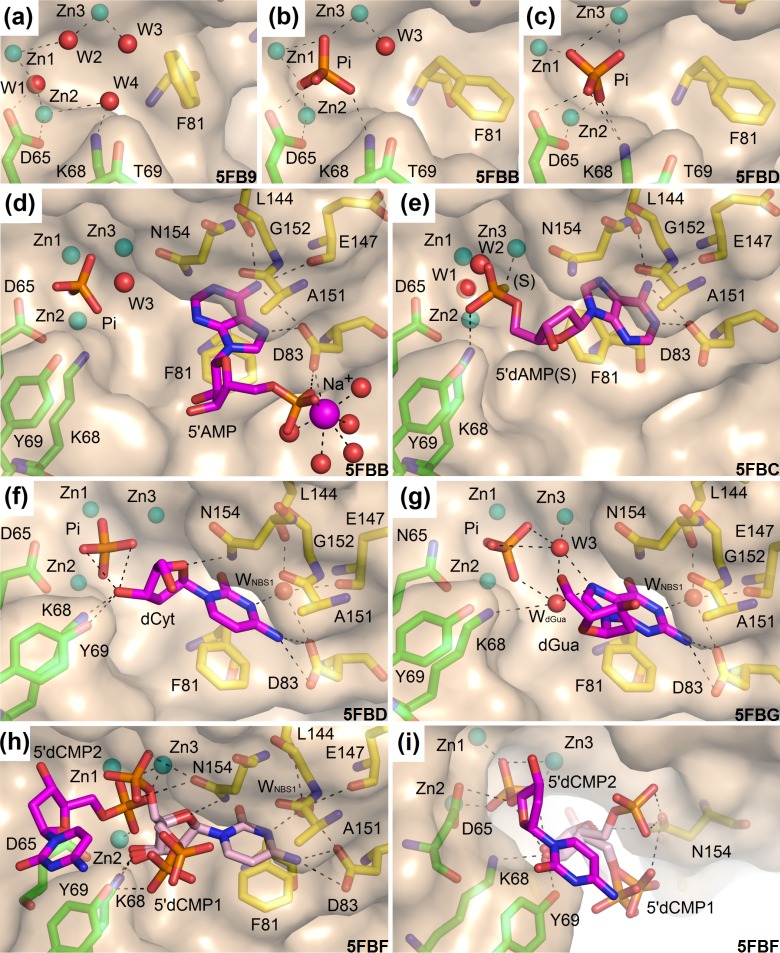
Fig 3. Observed binding of the ligands in S1 nuclease structures.
The catalytic zinc ions are shown as light blue spheres. Asp65 (Asn65 in the case of the mutant) is shown in sticks (carbon–green). Other residues involved in the zinc cluster coordination are not shown. Lys68 and Tyr69 interact with ligands and are shown as sticks (carbon–green). Residues forming NBS1 are shown as sticks (carbon–yellow). Important water molecules are shown as red spheres and phosphate ions as orange/red sticks. Selected interactions are shown as black dashed lines. Molecular graphics were created using PyMOL (Schrödinger, LLC). PDB ID of each structure is shown. (a) Binding of water molecules in the unoccupied zinc cluster in the structure 5FB9 –unoccupied (pH 5.5). (b) The first binding mode of the phosphate ion in the structure 5FBB–inhibitors (pH 6.5). 5’AMP is excluded from the graphics for clarity. (c) The second binding mode of phosphate ion in the structure 5FBD–nucleotidase products (pH 4.2). All water molecules are displaced and none of the oxygen atoms of the phosphate ion occupies the original positions of water molecules. dCyt is excluded from the graphics for clarity. (d) The inverted deep binding mode of 5’AMP in the complex 5FBB–inhibitors (pH 6.5). Water WNBS1 is replaced by the inhibitor. The zinc cluster is occupied by a phosphate ion. (e) The binding mode of 2'–deoxyadenosine 5'–thio–monophosphate in the complex 5FBC–remodeled (pH 5.5). The thiophosphate moiety binds outside the zinc cluster interacting only with Zn3 using its sulfur atom (marked as S), with Lys68 by the oxygen atom and through a water network with Asn154. In this deep binding mode WNBS1 is replaced. Notice the same position of the adenine amino group, similar to the position described in (d), but an entirely inverted orientation of the base. NBS1 is remodeled to the extended form. (f) The observed binding mode of 2'–deoxycytidine (carbon–magenta) in the complex 5FBD–nucleotidase products. The cytosine moiety interacts with the protein using several types of interactions. Its π–conjugated system interacts with Phe81 and the peptide bond between Ala151 and Gly152. It has a direct polar interaction with the side chain of Asp83 and several water–mediated interactions, including involvement of water inside the NBS1 site. The 2’–deoxyribose moiety binds close to the zinc cluster and interacts directly with Lys68Nζ, Tyr69Oη, Asn154Nδ2, and the phosphate ion. A similar binding occurs in the complex 5FBF–nuclease products (for the dCyt moiety of 5’dCMP) and in the complex 5FBG–mutant with products. (g) The observed binding mode of 2’–deoxyguanosine (carbon–magenta) in the complex 5FBG–mutant with products, chain B. Binding of the pyrimidine–like part of guanine mimics the orientation of cytosine inside NBS1 as shown in panel (f). N7 of the imidazole–like part is involved in the water network (W3 and WdGua) connecting this atom to Zn3, the phosphate ion, and Lys68Nζ. Asp154Nδ2 can interact with the π–conjugated electrons of the guanine moiety. (h) The observed binding mode of two molecules of 5’dCMP in the complex 5FBF–nuclease products (pH 4.2). The binding mode of the first 5’dCMP (carbon–pale pink) is almost identical with binding of 2’–deoxycytidine in the case of the complex 5FBD–nucleotidase products (panel f). The phosphate moiety is disordered and interacts either with Asn154 or with Lys68 and Tyr69. (i) The phosphate moiety of the second 5’dCMP in the complex 5FBF–nuclease products (carbon–magenta) binds in the zinc cluster in the second binding mode (as phosphate ion in 5FBD–nucleotidase products, panel f). The cytosine moiety likely interacts with Tyr69Oη (hydroxyl group) through its π–conjugated system.