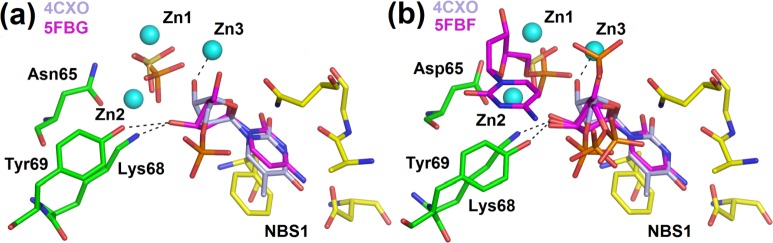
Fig 7. Two types of interactions of the O3’ oxygen with the active site in the S1–P1 nuclease family.
Zinc ions are shown as light blue spheres. Asp65 (Asn65 in the case of the mutant) is shown in sticks (carbon–green). Other residues involved in the zinc cluster coordination are not shown. Lys68 and Tyr69 interact with ligands and are shown as sticks (carbon–green). Residues forming NBS1 are shown as sticks (carbon–yellow). Phosphate ion is shown as orange/red sticks, sulfate ion as yellow/red sticks, and selected interactions as black dashed lines. Molecular graphics were created using PyMOL (Schrödinger, LLC). (a) Comparison of the deoxyribose binding and position of its O3’ in the structure 5FBG–mutant with products with its position in the structure of AtBFN2 (PDB ID: 4CXO [14]). The zinc cluster, residues involved in the interactions and NBS1 are shown only for S1 nuclease. Ligands present in the structure of S1 nuclease are phosphate and dCyt (carbon–magenta). Ligands present in AtBFN2 are sulfate and thymidine 5’–monophosphate (carbon–silver). Notice the difference in the positions of the O3’ oxygens and of the phosphate ion in S1 –excluding the interaction of O3’ with Zn3. (b) Comparison of deoxyribose binding and the position of its O3’ in the structures 5FBF–nuclease products and AtBFN2 (PDB ID: 4CXO [14]). The zinc cluster, residues involved in the interactions and NBS1 are shown only for S1 nuclease. Ligands present in the structure of S1 nuclease are two molecules of 5’dCMP (carbon–magenta). Ligands of AtBFN2 are displayed as in panel (a). Notice the difference in the positions of the O3’ oxygens and of the phosphate moiety in the case of S1 which excludes binding of O3’ to Zn3.