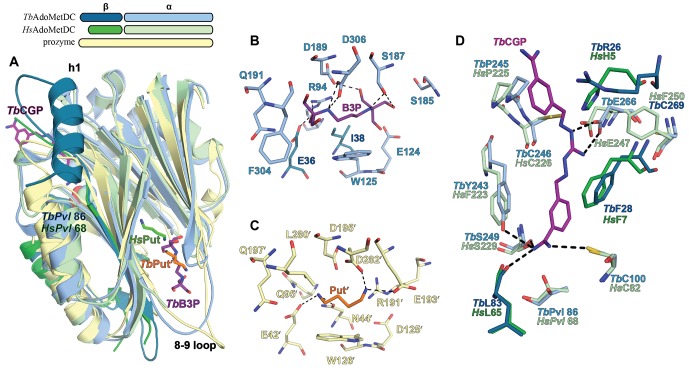
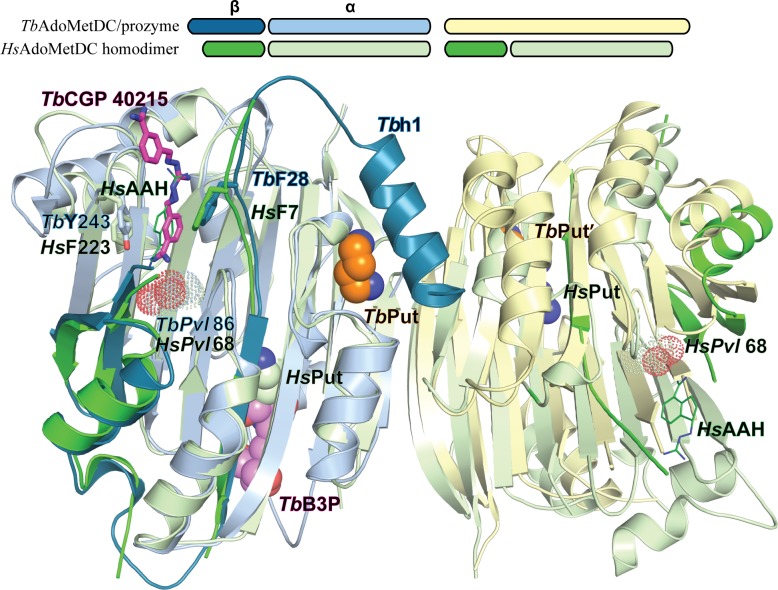
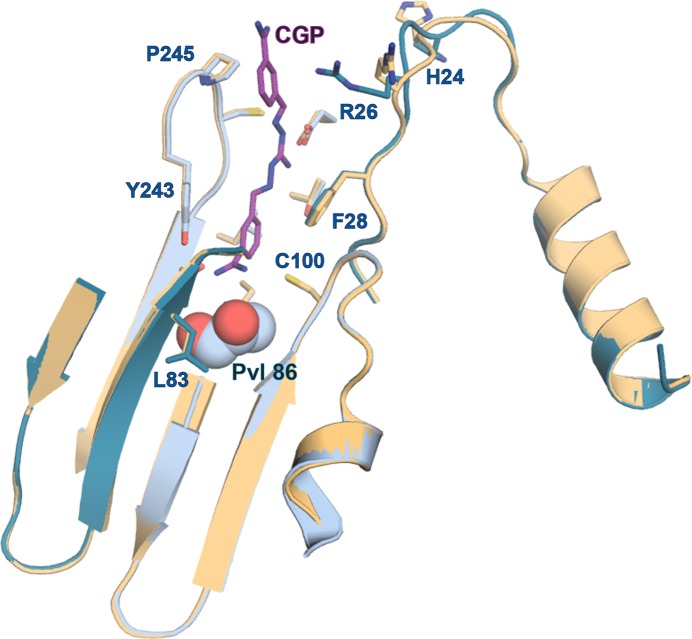
Figure 6. Ligand-binding sites in the CGP-TbAdoMetDC/prozyme heterodimer structure.
(A) Overlay of CGP-TbAdoMetDC β/α (teal/sky blue, CGP 40215 (TbCGP) and B3P in purple), prozyme (yellow, TbPut’ in orange) and HsAdoMetDC (3DZ6) β/α (dark green/light green, HsPut in green) structures viewed from the dimer interface. The schematic above the figure depicts color codes for the various chains. (B–C) Limited 4 Å shell showing the B3P- and putrescine-binding sites in TbAdoMetDC (B) and prozyme (C). (D) Overlay of the TbAdoMetDC CGP-binding site with HsAdoMetDC showing select residues in the 4 Å shell. H-bond interactions (distance < 3.3 Å) are shown by dashed lines. The electron density supporting ligand placement is shown in Figure 3—figure supplement 2, the complete structural alignment of Hs and Tb AdoMetDCs in Figure 6—figure supplement 1 and the comparison of the unliganded and liganded TbAdoMetDC structures in Figure 6—figure supplement 2.