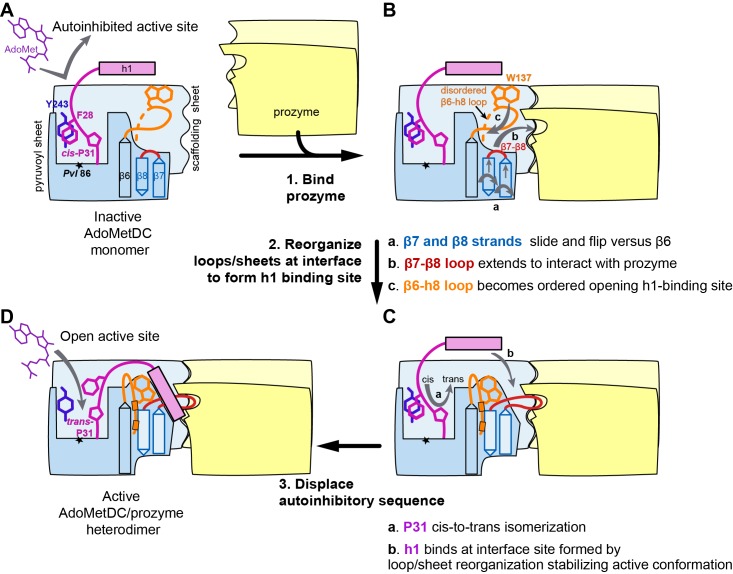
Figure 8. Mechanistic model of prozyme-induced TbAdoMetDC enzyme activation.
The model depicts a logical step-wise process assuming that the formation of the h1 binding site precedes insertion of the helix into the interface, but current data do not distinguish between a sequential versus a concerted activation mechanism and the ordering of events is hypothetical. (A) The inactive TbAdoMetDC monomer is composed of two β-sheets: pyruvoyl (blue) and scaffolding (light blue). The active site pyruvoyl residue (Pvl86, star) is blocked by the inhibitory sequence (S27–G30), which is oriented into the active site by the cis configuration of P31. This autoinhibitory closed confirmation is stabilized by π-π stacking between F28 (purple) and Y243 (blue). Residues N-terminal of S27, including helix h1 (purple rectangle), were not present in the monomer construct and their position in the diagram is hypothetical. (B) Binding of prozyme (yellow/light yellow) to AdoMetDC is nucleated by formation of the H-bond network between the two scaffolding sheets leading to formation of a continuous inter-subunit β-sheet that serves as a platform for the following conformational changes: (a) slipping of the interface strands β7 and β8 relative to β6, which results in flipping of their side chains from one surface to the other; (b) repositioning and elongation of the β7-β8 loop that forms the back of the h1 binding site, stabilized in this confirmation by interaction across the interface with prozyme; and (c) disordered-to-ordered transition and movement of the β6-h8 loop (H130-L144, orange) that leads to formation of the h1 binding pocket. (C) Upon the formation of the h1 binding pocket, (a) cis-to-trans isomerization of P31 displaces the autoinhibitory sequence from the active site and the open active site conformation is stabilized by (b) docking of the h1 helix at the dimer interface. (D) The active TbAdoMetDC/prozyme dimer is capable of binding ligands in the open active site, which leads to ~1000 fold increase in rates of AdoMet decarboxylation.